Copyright Notice
Copyright 2004-2024 Takashi Tanaka
This software is free for use, however, the author retains the copyright to this software. It may be distributed in its entirety only and may not be included in any other product or be distributed as part of any commercial software.
This software is distributed with NO WARRANTY OF ANY KIND. Use at your own risk. The author is not responsible for any damage done by using this software and no compensation is made for it.
This software has been developed, improved and maintained as voluntary work of the author. Even if problems and bugs are found, the author is not responsible for improvement of them or version up of the software.
If you are submitting articles to scientific journals with the results obtained by using this software, please cite the relevant references. For details, refer to Introduction.
Introduction
This document describes the instruction to use the free software SIMPLEX, a free electron laser simulation (FEL) code, and is located in "[SIMPLEX Home]/help", where "[SIMPLEX Home]" is the directory where SIMPLEX has been installed. Brief explanations on the software and numerical implementation of FEL equations are given here, together with a simple instruction of how to get started. Note that "MathJax" javascript library is needed to correctly display the mathematical formulas, which is available online. If you need to read this document offline, "MathJax" should be installed in "[SIMPLEX Home]/help" directory.
Overview
SIMPLEX, which stands for SIMulator & Postprocessor for free electron Laser EXperiments, is a computer program to simulate the amplification process of free electron lasers (FELs) and evaluate the light source performances, such as the radiation power growth, electron motion in the phase space, evolution of angular and spatial profiles of radiation field, etc. In order to perform the FEL simulations, a lot of parameters are required to specify the electron beam, lattice function, undulator, and numerical & boundary conditions. SIMPLEX is equipped with a graphical user interface (GUI), which helps the user to input these parameters. Possible errors in the undulator such as the magnetic field and trajectory errors can also be handled in the GUI.
Besides the above standard functions, the GUI automatically computes and shows relevant parameters related to the FEL system, such as the Pierce parameter, 1D & 3D gain lengths, and saturation power, so that the user can quickly check if their parameters are realistic in advance of actually starting the simulation. Light source characteristics, such as the lasing wavelength, bandwidth, source size and brilliance, are also estimated based on an assumption that the FEL radiation is spatially coherent.
In addition to pre-processing mentioned above, SIMPLEX offers a GUI-based postprocessor to facilitate retrieving desired information from the simulation results. For example, light source characteristics of FEL radiation can be retrieved from the radiation field profile, and the electron motion can be retrieved from the numerical data on macroparticles modeling the electron beam.
The numerical part of SIMPLEX ("solver") is written in C++11 with the standard template library (STL), which solves the equations describing the FEL process in the 3-dimensional (3-D) form. As explained in the next section, the simulations can be classified into two types according to how to deal with the slippage between electrons and radiation emitted by them. For details of numerical implementation, refer to [1].
In ver. 3.0, the solver has been widely revised to be consistent with the C++11 standard and to facilitate the maintenance and bug fix. In addition, numerical implementation of FEL equations has been improved to be more CPU-efficient and to reduce the simulation time. In addition to the above revisions, two important upgrades have been made. First, the format of the input parameter file has been changed from the original (and thus not readable in other applications) one to JSON (JavaScript Object Notation) format. Because the output data is also given by a JSON file, it is now easy to communicate with other 3rd-party applications.
Simulation Modes
In order to investigate the FEL amplification process, it is necessary to solve three equations: (1) equation of motion to describe the motion of each electron in the phase 6-D (x,x',y,y',t,γ) space, (2) energy equation to describe the energy exchange between electrons and radiation, and (3) wave equation to describe the evolution of radiation field. Let us call these equations "FEL equations" hereinafter. The main function of an FEL simulator is to numerically solve the FEL equations.
The wave equation describes evolution of the electric field of radiation and is given by \[\left[\nabla^2+2i\frac{\omega}{c}\left(\frac{\partial}{\partial z}+\frac{1}{c}\frac{\partial}{\partial t}\right)\right]\mathbf{E}\mbox{e}^{-i\omega(z/c-t)}+\mbox{c.c.}=\mu_0\frac{\partial\mathbf{j}}{\partial t}+\frac{\nabla\rho}{\varepsilon_0},\] where E is the complex amplitude of radiation field, ω is the frequency of radiation, j and ρ are the current and charge densities of the electron beam.
Assuming that the electron beam is infinitely long with constant beam parameters such as the emittance, energy, current, and offset (angular and positional), and also the radiation is completely monochromatic, the partial differentiation with respect to time can be dropped from the wave equation. In such a case, the simulation algorithm is considerably simplified and the simulation is referred to as "steady-state".
If the above assumption cannot be applied, the wave equation should be solved with the time dependence taken into account, namely, the slippage between the electron beam and radiation should be considered. To be specific, the radiation emitted by the electrons slips out of them by the so-called slippage length, which equals the fundamental wavelength of undulator radiation. The FEL simulation that takes into account the slippage effect is referred to as "time-dependent", which requires a lot of computation time because the algorithm to solve the FEL equations should be applied over the whole electron bunch.
In order to roughly estimate the FEL performance such as the saturation power, saturation length, radiation spatial profile and motion of the electrons in the 6-D phase space, the steady-state simulation suffices. On the other hand, the time-dependent simulation should be performed for various purposes, e.g., the start-to-end simulations for SASE (self-amplified spontaneous emis-sion) FELs, and investigations of the effects due to the wakefield induced in the undulator line.
Functions Available
Besides the normal FEL simulation procedures described in the preceding section, SIMPLEX offers various functions to investigate the performance of FEL.
Rough Estimation of FEL Performances
A number of FEL parameters and light source characteristics can be roughly estimated by an-alytical formulae instead of rigorous simulations. SIMPLEX preprocessor automatically computes these values with the given conditions, in order to help the users not only to check if their pa-rameters are valid but also to understand the meanings of parameters.
Electron Beam Data Manipulation
In order to perform the so-called start-to-end simulation, initial conditions of the electron beam injected to the undulator should be specified. SIMPLEX accepts the user input by importing a file that stores the numerical data of the sliced beam parameters along the bunch. Another way is to load the macroparticle distribution in the 6-D phase space, which is probably generated by an external program. In addition, the preprocessor offers a function to analyze the macroparticle distribution to obtain the sliced beam parameters such as the current, emittance, energy spread, and so on.
Modeling the Undulator Field Error
In normal FEL simulations, the undulator magnetic field is assumed to be completely sinusoidal, which is not the case for the real undulator. SIMPLEX can import the magnetic field distribution data actually measured with a field measurement apparatus such as Hall-effect sensors. Other possible errors of the magnetic field, such as the geomagnetic field and demagnetization due to radiation damage, can be investigated by importing the field-distribution data including these effects. In addition to the above function to specify the undulator error, SIMPLEX offers an option to automatically generate the field errors which reproduce the phase error and trajectory wander in respective undulator segments.
Modeling the Alignment Error
Besides the magnetic field error, there is another error source in the undulator line: alignment errors such as the discrepancy in the gap value between undulator segments, tilt and offset in the vertical position of each undulator, the trajectory error due to misalignment of beam position monitors, and so on. SIMPLEX can perform FEL simulations with all these misalignment effects taken into account.
Wakefield Effects
The wakefield is induced by interaction between the electron and surrounding environment such as the beam pipe, and induces correlated energy variation in the electron bunch. In order to investigate the wakefield effects on the FEL amplification process, it is required to specify the profile of the wakefield along the electron bunch. In SIMPLEX, the wakefield can be computed internally using analytical expressions derived in several research papers. It is also possible to specify the wakefield distribution by importing the wakefield data.
Configuration of Lattice Functions
In general, there exists an optimum betatron function for a given set of electron-beam and undulator parameters and thus the layout and strength of focusing magnets in the undulator line should be designed so that the average betatron function is optimized. SIMPLEX offers a couple of focusing magnet layouts that are periodically arranged with the undulator segments, which are supposed to be regularly spaced. SIMPLEX preprocessor offers an option to compute the optimum initial Twiss parameters for such a periodic lattice.
Post-Processing
The raw data of simulation results of SIMPLEX, i.e., the complex amplitude of radiation field and macro-particle distribution at each slice are saved in binary files. In order to process them and visualize the results (post-processing), such as the spectrum, instantaneous radiation power, and bunch factor, a GUI-based post-processor is available.
Getting Started
- Open a parameter file by running [File]-[Open a Parameter File] command.
- Edit or input parameters in the GUI window. All the parameters and configurations are categorized into several categories, and are shown in a box titled with each category name. To change the category shown in the GUI, select a relevant tab window.
- Run [Run]-[Start Simulation] command to start a simulation with current parameters.
- A "Progressbar" appears to inform the simulation status.
- The simulation results are saved in files specified by the user with different suffices. For example, the radiation power and bunch factor averaged over the whole bunch are saved in a file with a suffix "json" in the JSON format, together with the input parameters used in the simulation
- To verify the simulation results, click "Post-Processing" tab, select the name of the output JSON file, and item(s) to check for visualization. Refer to Post-Processing for details.
Operation of the GUI
Tab Panels
SIMPLEX GUI is composed of 6 tab panels entitled as Electron Beam & Seed, Undulator Line, Options, Simulation Controls, Pre-Processing, and Post-Processing. Note that FEL Performance and Output File subpanels are shown when one of "Electron Beam & Seed", "Undulator Line", "Options" and "Simulation Controls" tab panels is selected.
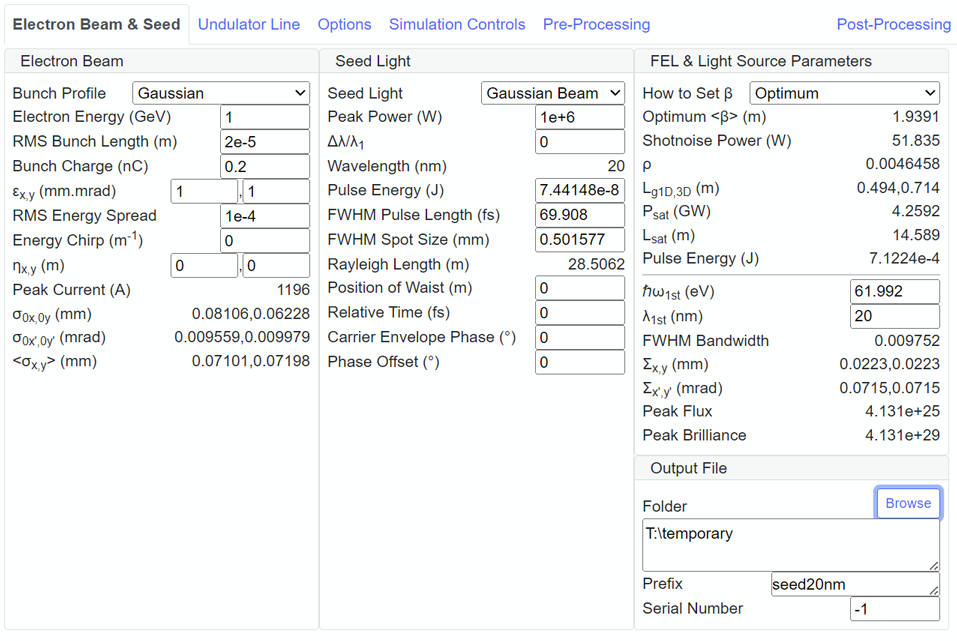
| Title | Operation | Related Subpanels |
| Electron Beam & Seed | Display and edit the parameters and configurations related to the electron beam, seed light, and importing the former simulation result. | Electron Beam, Seed Light and SIMPLEX Output |
| Undulator Line | Display and edit the parameters and configurations related to the undulator and lattice functions. | Undulator and Lattice |
| Options | Display and edit the parameters and configurations related to the alitmnent tolerance, wakefield in the undulator, chicane to be inserted in the undulator line, and trajectory error and sources to spoil the overlap between the electron beam and seed light. | Alignment, Wakefield, Chicane and Dispersion |
| Simulation Controls | Display and edit the parameters and configurations related to the simulation conditions and data save. | Simulation Conditions and Data Dump Configurations |
| Pre-Processing | Assist the pre-processing, i.e., visualization and configuration of the simulation conditions, which are not displayed in the subpanels explained above. | Pre-Processing |
| Post-Processing | Assist the post-processing, i.e., visualization of the simulation results. | Post-Processing |
Menu Commands
Menu commands available in SIMPLEX are described below.
File
| Menu | Details |
| Open a Parameter File | Open a SIMPLEX parameter file. Although those for older (≦ 2.1) versions are automatically converted to the new format, the user is strongly recommended to check if the conversion is OK. |
| Save | Save all the parameters and options in the current file. |
| Save As | Save all the parameters and options in a new file. |
| Load Output File | Import the former simulation results for visualization. |
| Load Post-Processed Result | Import the former post-processred results for visualization |
| Exit | Quit SIMPLEX and Exit |
Run
| Menu | Details |
| Create a New Process | Create a simulation process with the current parameters and options. |
| Export Simulation Settings | Export the current parameters and options to a file, or download it (web-application mode), which can be used as an input file to directly call the solver. |
| Start Simulation | Start a new simulation, or launch the simulation process. |
Help
Open the reference manual or show the information about SIMPLEX.
Edit the Plot
Besides standard Plotly.js configurations, a number of options to edit the graphical plot in the post- and pre-processors are available. To do so, click the small icon located in the top-right side of the plot. Then a dialog box pops up to let the user edit the plot in the following configurations.
| Item | Details | Available Options |
| Scale | Select how to normalize the animation plot | For Each: y-/z-axis scale is normalized by the maximum value for each slide |
| By Maximum: y-/z-axis scale is normalized by the maximum value over the whole slides | ||
| X-axis Scale | Select the scale for x axis | Linear, Logarithmic |
| Y-axis Scale | Select the scale for y axis | Linear, Logarithmic |
| Plot Type | Select the type of the 1D plot | Line, Line & Symbol, Symbol |
| Symbol Size | Size of the symbol | Input a number |
| Line Width | Width of the line | Input a number |
| 2D Plot Type | Select the type of the 2D plot | Contour: contour plot with a specific color map |
| Surface (Color Scale): surface plot painted with a specific color map | ||
| Surface (Shading): shaded surface plot illuminated by a specific light source | ||
| Color | Select the color of the light source to create a shaded surface plot | Select from the color picker dialog. |
| Color Map | Select the color map. Several built-in options are available but cannot be customized | Default, Rainbow, Blackbody, Earth, Greys |
| Wireframe | If checked, grid lines are shown on the surface plot |
Simulation Setup
Details of how to setup and start the simulations are presented here.
General Method
Open a Parameter File
Upon being started, SIMPLEX tries to load parameters from the parameter file that was opened last time. If successful, the parameters are shown in the GUI. If SIMPLEX is run for the first time after installation, default parameters will be shown. To open a new SIMPLEX parameter file, run [File]-[Open a Parameter File] command. In the initial setting, the parameter files are found in the directory "[SIMPLEX Home]/prm" with a default suffix "json", where "[SIMPLEX Home]" is the directory in which SIMPLEX has been installed.
Rough Estimation
Before starting a simulation, the user is recommended to roughly estimate the expected performances of the FEL system, such as the gain length, saturation power, lasing wavelength, brilliance, etc., to make sure that the input parameters are not far from reasonable ones. In SIMPLEX, these values to represent the FEL performance are automatically evaluated using analytical and empirical expressions on FEL physics [2]-[4] and shown in the FEL Performance subpanel. Note that these values are approximate ones and are given just for reference.
Arrange the Output File
Arrange the output JSON file to save the simulation results in the Output File subpanel. SIMPLEX generates at most 3 files (or more, dependenig on the maximum harmonic number) to save the simulation result with the names of *.json, *.par, *.fld, where * stands for the data name specified by the user. Refer to Output File about how the data name is given.
The gain curve (growth of the pulse energy etc. along the undulator line) is saved in the "*.json" file (hereafter, "output JSON file") together with the input parameters and configurations. In addition, temporal, spectral, spatial and angular profiles of radiation are optionally saved if specified. For details, refer to Data Dump Configurations. Note that the data is saved in the JSON Format. The output JSON file can be loaded for visualization of the gain curve and radiation profiles, or for Raw Data Processing. Besides such post-processing operations, it can be loaded as a parameter file for another simulation.
The radiation and particle data (raw data) are saved in the "*.fld" and "*.par" files, respectively, in a binary format. The raw data in these files will be used in another simulation, or post-processed later with the assistance of the output JSON file as mentioned above.
Start Simulation
Run [Run]-[Start Simulation] command to start a single simulation. Then "Create a New Process" subpanel is displayed to indicate the progress of simulation. To cancel the simulation, click "Cancel" button.
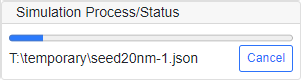
Note that the serial number is automatically incremented once the simulation is started, unless it is not negative (-1). This is to avoid the overlap of data names in performing successive simulations. When the simulation is completed, the "Create a New Process" subpanel vanishes and the result is imported in the "Post-Processing" panel for visualization.
Verify the Result
Upon completion of a simulation, the output JSON file is automatically loaded and the gain curve (pulse energy vs. longitudinal position) is plotted in the "Post-Processing" subpanel to quickly view the results. Refer to Post-Processing for details about how to operate the Post-Processing.
Create a New Process
To configure a number of simulations with different conditions, run [Run]-[Create Process] command every time you finish specifying all the parameters. Then the "Create a New Process" subpanel appears to show the simulation list currently saved in a temporary memory. Repeat the above porcess until all the simulations are specified. Click "Remove" button to delete the selected process, or "Cancel All" to clear out all the processes. Run [Run]-[Start Simulation] command to start the simulation processes, then a progressbar is displayed to show the status of each process as shown below.
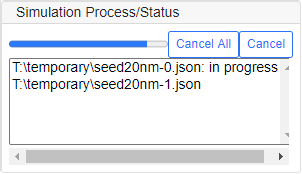
Scanning a Parameter
Besides the method described above, it is possible to configure a lot of processes at once by scanning a specific parameter. To do so, right click the target parameter in one of the subpanels, and click [Scan This Parameter] in the context menu. Then specify the configurations for scanning in the dialog box as shown below. Note that the context menu does not pop up for parameters that cannot be used for scanning.
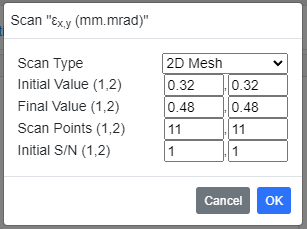
Input the initial & final values, and number of points for scanning. For several parameters to be specified by an integer, scanning interval instead of the number of points should be given. If the target parameter forms a pair, such as εx, y (horizontal and vertical normalized emittances), the user is requested to select "Scan Type". "1D: Single" means that only the target parameter (εx or εy in this example) is varied from the inital to the final values. "1D: Link 1st/2nd" means that both of the pair are simultaneously varied. In contrast to these 1D scan types, "2D Mesh" varies the both parameters in a 2D manner, and thus N1N2 processes are created, where N1,2 stands for "Scan Points (1,2)" parameters.
After configuration, click "OK" button. Then the specified parameters are saved in a temporary memory and the scanning process is saved in the simulation list. Run [Run]-[Start Simulation] command to start the simulation. Note that a serial number starting from "Initial S/N" is attached to the data name to avoid overlap. If "2D Mesh" is chosen, the serial number is given like "_m_n", where m and n mean the serial numbers corresponding to the 1st and 2nd parameters of the pair.
Parameter List
All the parameters and configurations needed to perform an FEL simulation are classified into several categories in SIMPLEX, and can be edited in the subpanels tiltled with the category name. Details of them are explained in the followings for each category.
Electron Beam
The parameters and configurations in the "Electron Beam" subpanel are related to the specifications of the electron beam. Details are summarized below.
| Parameter/Option | Detail |
| Bunch Profile | Bunch profile of the electron beam in the 6D phase space. For details, refer to Bunch Profile. |
| Particle Distribution | Path to a local file containing the particle data. Input the absolute path in the text box, or click "Browse" and specify the file in the dialog box. Note that the data file should be an ASCII file, and each line is composed of 6 values specifying the position of a single macroparticle in the 6D phase space. After specifying the data, the user is strongly recommended to perform "Analyze Particle Data" for configuration of the particle data such as the units and column indices for respective coordinates. |
| Electron Energy (GeV) | Slice (representative) energy of the electron beam. |
| RMS Bunch Length (m) | RMS bunch length of the electron beam. |
| Bunch Length (m) | Full bunch length of the electron beam. Available if "Boxcar" is chosen for the electron bunch profile. |
| Bunch Charge (nC) | Bunch charge of the electron beam. |
| εx,y (mm.mrad) | Normalized emittance of the electron beam. |
| RMS Energy Spread | Energy spread and chirp of the electron beam. |
| Energy Chirp (m-1) | |
| ηx,y (m) | Dispersion functions of the electron beam. |
| Peak Current (A) | Peak current of the electron beam evaluated from relevant parameters. |
| σ0x,0y (mm) | Beam size and angular divergence at the undulator entrance. |
| σ0x',0y' (mrad) | |
| R56 (m) | Strength of the virtual dispersive section located in front of the undulator. Available if "SIMPLEX Output" is chosen for the electron bunch profile, and may be needed to simulate the HGHG/EEHG FELs. Refer to Configuration to Use the Former Result for details. |
Bunch Profile
The distribution functions of the electron beam assumed in the FEL simulation, or the profiles in the 6D phase space available in SIMPLEX, are summarized below. Note that the custom data needed for several types can be imported in the Import Custom Data tab panel.
| Type | Detail |
| Gaussian | Distribution function in the 6D phase space is given by a 6D Gaussian function. |
| Boxcar | Same as "Gaussian", but the current profile is given by a boxcar function. |
| SIMPLEX Output | Macroparticles are loaded from the former simulation result. To specify the former simulation, refer to SIMPLEX Output. |
| Current Profile | Same as "Gaussian", but the current profile is given by custom data. |
| Slice Parameters | Same as "Current Profile", but the slice parameters are given by custom data. |
| E-t Profile | Same as "Gaussian", but the profile in the (E-t) phase space is given by custom data. |
| Particle Distribution | Beamlets are loaded from a data file that contains the particle distribution in the 6D phase space. The user should specify the path of the data in "Particle Distribution" and specify its format. Refer to Analyze Particle Data for details of how to specify the data format and how to verify its validity. |
Seed Light
The parameters and configurations in the "Seed Light" subpanel are related to the specifications of the electron beam. Details are summarized below.
| Parameter/Option | Detail |
| Seed Light | Type of the seed light. For details, refer to Seed Type. |
| Peak Power (W) | Peak power of the seed light. |
| Δλ/λ1 | Wavelength of the seed light relative to the undulator fundamental wavelength. |
| Wavelength (nm) | Wavelength of the seed light. |
| Pulse Energy (J) | Pulse energy and pulse length of the seed light. |
| FWHM Pulse Length (fs) | |
| FWHM Spot Size (mm) | Source size and Rayleigh length of the seed light. |
| Rayleigh Length (m) | |
| Position of Waist (m) | Longitudinal position where the seed light forms a beam waist |
| Carrier Envelope Phase (°) | Carrier envelope phase, group delay dispersion and third order dispersion of the seed light. |
| GDD (fs2) | |
| TOD (fs3) |
Seed Type
Types of the Seed light available in SIMPLEX are summarized below.
| Type | Detail |
| N.A. | No seed light is supposed. |
| Gaussian Beam | Transform-limited Gaussian beam is supposed. |
| Chirp Pulse | Chirped Gaussian beam is supposed. |
| SIMPLEX Output | Radiation profiles are loaded from the former simulation result. |
SIMPLEX Output
The results of the former simulation, namely, the distribution of the macripartiles and radiation profiles can be reused in another simulation. To enable this option, related raw data should be exported in the former simulation (Data Dump Configurations).
| Parameter/Option | Detail |
| Data Name | Path to the output data file |
| Step Index to Retrieve Data | Step index to retrieve the raw data of the former simulation, counted from the final step. For example, "-1" means that the raw data at the (N-1)-th step are used. |
| z (m) | Longitudinal position corresponding to the above index. |
| undefined | Length of the drift section between the exit of the former simulation and the entrance of the current one. |
| Photon Energy (eV) | Fundamental energy used in the former simulation |
Configuration to Use the Former Result
To use the former simulation result as
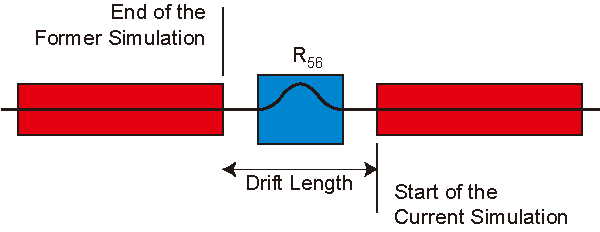
Undulator
The parameters and configurations in the "Undulator" subpanel are related to the specifications of the undulator. Details are summarized below.
| Parameter/Option | Detail |
| Main Parameters | |
| Undulator Type | Undulator type. For details, refer to Undulator Type. |
| K Value | Deflection parameter (K value) of the undulator. Note that K⊥ is defined as \[K_{\perp}=\sqrt{K_x^2+K_y^2}\] |
| K⊥ | |
| tan-1(Kx/Ky) (°) | Specifies the ratio of the horizontal and vertical K values. |
| Multi-Harmonic Contents | Arrange the harmonic components of "Multi-Harmonic". For details, refer to Multi-Harmonic. |
| λu (mm) | Magnetic Period Length of the undulator |
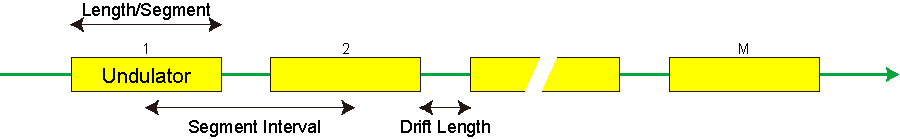
| Length/Segment (m) | Parameters to specify the undulator line, which is supposed to be composed of several segments and drift sections in between. For details, refer to Undulator Line Parameters. |
| Number of Segments | |
| Segment Interval | |
| Peak Field (T) | Peak field of the undulator evaluate from the K value |
| Periods/Segment | Number of undulator periods/segment |
| Slippage in Drift (2π) | Number of slippage in the drift section |
| Extra Slippage (°) | Extra slippage in the drift section |
| Tapering | Type of undulator taper. For details, refer to Taper Type. |
| Undulator Model | Type of undulator model. For details, refer to Undulator Models. |
| Undulator Taper Parameters | |
| Optimization | Specifies how to optimize the taper rate. For details, refer to Automatic Taper Optimization. |
| Target Slice Position (m) | Target slice position for taper optimization |
| Initial Segment | Four parameters to specify the undulator taper. For details, refer to How to Setup Tapering. |
| Increment Segment Interval | |
| Base Linear Taper (m-1) | |
| Taper Increment (m-1) | |
| K Value@Exit | K value at the exit of the undulator |
| Resonance Detuning@Exit | Detuning at the exit of the undulator |
| Undulator Model Configurations | |
| Random Number Auto Seeding | If enabled, seed number for the random number generator is automatically determined |
| Random Number Seed | Seed number for the random number generator to model the undulator error |
| σφ (°) | RMS phase error |
| σB/B (%) | RMS field deviation |
| σx,y (mm) | RMS trajectory error |
| Apply to All Segments | Apply the error to all the segments |
| undefined | Target segment number to apply the error |
Undulator Type
Undulator types available in SIMPLEX are summarized below.
| Type | Details | Radiation Field for Simulation |
| Linear | Conventional linear undualtor with a vertical magnetic filed to generate 100% linearly polarized radiation. | Scalar: Ex |
| Helical | Helical undulator to generate 100% circularly polarized radiation. | Scalar: Ex+iEy |
| Elliptical | Elliptical undulator to generate elliptically polarized radiation. | Vector: (Ex, Ey) |
| Multi-Harmonic | Special undulator generating a magnetic field composed of multiple harmonic components. | Vector: (Ex, Ey) |
Undulator Line Parameters
The undulator line is supposed to be composed of a number of undulator segments and drift sections in between, where quadrupole magnets are installed (FODO etc. are selected for the Lattice Type).
Multi-Harmonic
This is an undulator scheme generating a magnetic field composed of multiple harmonic components, whose configuration is specified in the spreadsheed titled Multi-Harmonic Contents. Kx Ratio ($=R_n$) and Phase ($=P_n$, in degrees) refer to the fraction and phase of the horizontal field of the n-th harmonic component, where n is the number indicated in the 1st column.
The K value corresponding to the n-th harmonic is defined as \[K_{xn}=K_x\frac{R_n}{\sqrt{R_1^2+R_2^2+\cdots+R_N^2}},\] where $K_x$ is the (total) K value and $N$ is the maximum harmonic number.
The field distribution is defined as \[B_{x}(z)=\sum_{n=1}^{N}B_{xn}\sin\left[2\pi\left(\frac{nz}{\lambda_u}+\frac{P_n}{360}\right)\right],\] where $B_{xn}$ is the peak field corresponding to the K value of the n-th harmonic.
Similar definitions of $K_{yn}$ and $B_{y}(z)$ for the horizontal field componeents.
Taper Type
Tapering options available in SIMPLEX are summarized below.
| Type | Details |
| N.A. | Tapering is not applied. |
| Stair-Like | Each undulator segment cannot be tapered; the K value is constant within a single segment and jumps at the entrance of the next segment. |
| Continuous | Each undulator segment can be tapered; the K value changes gradually from the entrance to the exit in a single segment, and thus the K value varies continuously along the undulator. |
| Custom | Customize the K value offset and slope for each segment. |
Custom Taper
Customize the K value and its gradient along the longitudinal axis for each undualtor segment, as defined below.
\[K=K_0+\Delta K+\frac{dK}{dz}(z-z_{mid})\]
where $K_0$ is the nominal K value, $\Delta K$ is the offset, $dK/dz$ is the gradient, and $z_{mid}$ is the longitudinal coordinate of the target segment.
Undulator Models
Undulator models available in SIMPLEX are summarized below.
| Type | Details |
| Ideal | Magnetic fields of all the undulator segments are ideal. |
| Specify Error | Specify the magnetic field errors. |
| Import Data | Use the field distribution given by custom data. To import the custom data, refer to Arrange Undulator Data. |
How to Setup Tapering
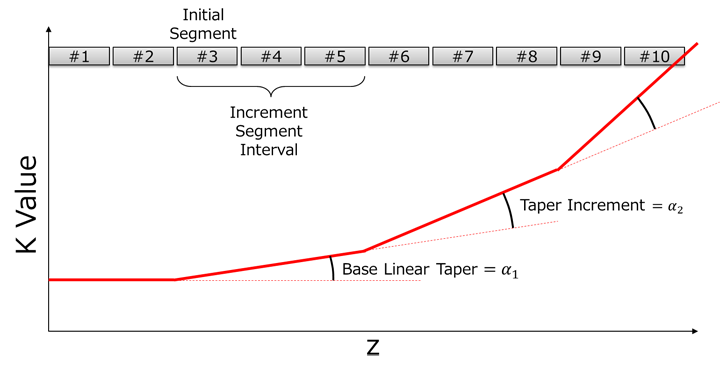
In order to specify the undulator taper, four parameters should be specified: "Initial Segment", "Increment Segment Interval", "Base Linear Taper" (α1), and "Taper Increment" (α2). The meanings of these parameters are schematically illustrated in the above figure, in which the K value along the undulator axis is plotted when both "Initial Segment" and "Increment Segment Interval" are assumed to be 3. In this example, the taper starts at the 3rd segment with the taper rate (dK/dz) of α1, which continues up to the 5th segment. Then the taper rate changes to α1+α2 at the 6th segment and continues up to the 8th segment, and again changes to α1+2α2 at the 9th segment. This kind of periodic increment continues up to the final undulator segment. The above taper configuration is to model the quadratic taper, and in fact is more convenient for the taper optimization than directly looking for the optimum coefficients of the 2nd order polynomial specifying the taper profile.
Automatic Taper Optimization
SIMPLEX offers a scheme to automatically adjust the taper rate to enhance the FEL output by a number of means as summarized below.
| Type | Details |
| N.A. | Optimization is not applied. |
| Compensate Wake | Besides what is specified by the user, additional tapering is applied to compensate for the energy loss by the wakefield evaluated at the target slice position. |
| Projection | The taper rate is automatically determined to compensate for the energy loss in the whole electron bunch. |
| Slice | The taper rate is automatically determined to compensate for the energy loss at the target slice position. |
Lattice
The parameters and configurations in the "Lattice" subpanel are related to the arrangement and specifications of the quadrupole magnets and initial Twiss parameters of the electron beam. Details are summarized below.
| Parameter/Option | Detail |
| Lattice Type | Type of the lattice. Refer to Lattice Type for details. |
| QF Gradient (T/m) | Field gradient of the quadrupole magnets. |
| QD Gradient (T/m) | |
| QF Length (m) | Length of the quadrupole magnets. |
| QD Length (m) | |
| QF-QD Distance (m) | Distance between the focusing and defocusing mangets. |
| # Periods/Segment | Number of FODO periods in a single undulator segment. |
| βx0,y0 (m) | Initial Twiss parameters at the undulator entrance. |
| αx0,y0 (m) | |
| Optimum β (m) | Optimmum betatron function evaluate to maximize the saturation power. |
Lattice Type
SIMPLEX offers 5 types of focusing magnet arrangements as shown below. Except for the "Undulator Combined" type, the focusing magnets are placed at the drift sections between undulator segments.
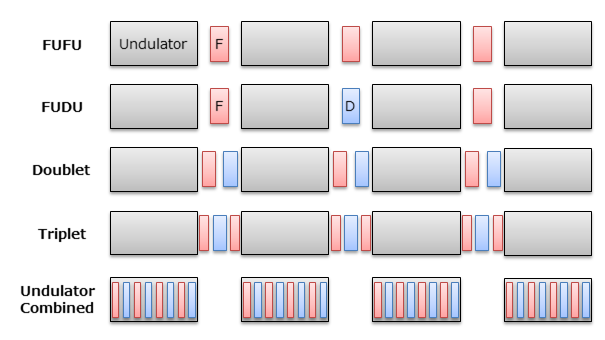
Alignment
The parameters and configurations in the "Alignment" subpanel are related to the alignment tolerance of the components in the undulator line; this is to evaluate the effects of the misalignment on the FEL amplification process. Details are summarized below.
| Parameter/Option | Detail |
| Undulator Alignment | Type of the undulator alignment error. For details, refer to How to Specify the Alignment Error. |
| ΔK Tolerance | RMS deviation of the K value |
| Slippage Tolerance (°) | RMS deviation of the phase slip |
| Offset for Each Segment | Specifies the offset of the K values and slippage for each segment |
| BPM Alignment | Type of the BPM alignment error. For details, refer to How to Specify the Alignment Error. |
| x,y Tolerance (mm) | Tolerance of the BPM alignment |
| Random Number Auto Seeding | If enabled, seed number for the random number generator is automatically determined |
| Random Number Seed | Seed number for the random number generator to model the alignment errors |
How to Specify the Alignment Error
Options to specify the alignment errors are summarized below.
| Type | Details |
| Ideal | All the components are aligned perfectly. |
| Specify Offset | K values and slippage values of target undulator segments are misaligned. |
| Specify Tolerance | Related components are aligned with a specific tolerance. |
Wakefield
The parameters and configurations in the "Wakefield" subpanel are related to the parameters and configurations related to the wakefield, which usually degrades the FEL gain.
Wakefield Evaluation
Wakefield is an electromagnetic wave induced by interaction between electrons and surrounding environment, and induces an energy modulation in the electron bunch, which eventually degrades the FEL gain. In SIMPLEX, several wakefield types can be taken into account to investitage their effects. Note that the wakefield is evaluated only once, before starting a simulation using the current profile at the undulator entrance, and is not modified during the simulation process. This means that the density modulation of the electron beam induced by the FEL interaction is not taken into accout for evaluating the wakefield.
- Resistive Wake; the resistive wakefield is evaluated using the expression derived in [5].
- Surface Roughness; the wakefield induced by the surface roughness can be in principle computed using the expressions derived in [6] and [7] with the knowledge of the surface profile. Instead of them, SIMPLEX uses modified expressions that denote the wakefield induced by the surface whose profile consists of randomly distributed bumps with a Gaussian spectrum.
- Dielectric Layer: if the surrounding conductor is covered with a thin dielectric layer due to, e.g., oxidation, wakefield is induced as discussed in [8].
- Space Charge: the space charge, or the "self-induced wakefield without any surrounding structure", is evaluated using the expressions derived in [9].
Details of the parameters and configurations are summarized below.
| Parameter/Option | Detail |
| Wakefield On | Enables the wakefield |
| Aperture (m) | Aperture of the surrounding beam duct |
| Resistive | Enables the resistive wall wakefield |
| Resistivity (Ωm) | Resistivity and relaxation time of the wall material |
| Relaxation Time (sec) | |
| Parallel Plate | If enabled, a parallel-plate configuration is used for the resistive-wall wakefield |
| Surface Roughness | Enables the wakefield induced by surface roughness |
| RMS Height (m) | Height and correlation length to specify the profile of the rough surface |
| Correlation Length (m) | |
| Dielectric Layer | Enables the wakefield induced by a dielectric layer |
| ε/ε0 | Permitivity and thickness of the dielectric layer |
| Thickness (m) | |
| Dielectric Layer | Enables the space charge effect |
| Additional Custom Wake | Enables the additional wakefield defined by custom data |
Chicane
The parameters and configurations in the "Chicane" subpanel are related to the chicane to be inserted in the undulator line. The details are summarized below.
| Parameter/Option | Detail |
| Chicane On | Replaces a single undulator segment by a magnetic chiane composed of 4 identical bending magnets. |
| Dipole Field (T) | Field strength, length, and distance of the bending magnets |
| Dipole Length | |
| Dipole Distance | |
| Beam Offset (mm) | Trajectory offset and time delay given by the chicane. |
| Electron Delay (fs) | |
| Chicane Position | Undulator segment number to be replace by the chicane. |
| Rearrange After Chicane | If enabled, the macroparticles are rearranged so that the microbunches induced before the chicane are smeared out. |
| Monochromator Type | Type of the monochromator to be inserted in the chicane for the self-seeding scheme [10][11]. |
| Crystal Type | Type of the crystal for the monochromator |
| ℏω (eV) | Photon energy of the monochromator |
| Bragg Angle (°) | Bragg angle for the monochromator energy |
| |Fg| | Form factor, lattice constant, and unit cell volume of the crystal |
| Lattice Spacing (nm) | |
| Unit Cell Volume (nm3) | |
| Bandwidth | Bandwidth of the crystal monochromator |
| Crystal Thickness (mm) | Thickness of the crystal |
| Relative Timing (fs) | Arrival time of radiation with respect to the electron beam after the chicane |
Dispersion
The parameters and configurations in the "Dispersion" subpanel are related to the injection errors and single kick; this is to evaluate the effects of the trajectory error and loss of overlap between the electron beam and seed light.
| Parameter/Option | Detail |
| e- Injection Error | Enables the injection error of the electron beam. |
| Δx,y (mm) | Positional and anglular offset of the electron beam at the undulator entrance. |
| Δx',y' (mrad) | |
| Single Kick | Adds a single kick at a certain position in the undulator line. |
| Kick Position (m) | Longitudinal position and angle of the single kick. |
| Kick Angle x,y (mrad) | |
| Seed Injection Error | Enables the injection error of the seed light. |
| Seed Δx,y (mm) | Positional and anglular offsets of the seed light at the undulator entrance. |
| Seed Δx',y' (mrad) |
Simulation Conditions
The parameters and configurations in the "Simulation Conditions" subpanel are related to the conditions to perform the FEL simulation. Detals are summarized below.
| Parameter/Option | Detail |
| Simulation Mode | Selects the simulation mode. Refer to Simulation Mode. |
| Simulation Option | Enables a special numerical algorithm to reduce the number of macropaticles needed to obtain a resliable simulation result. |
| Auto Integration Step | The longitudinal step to solver the FEL equation is automatically determined based on the approximate gain length. |
| Random Number Auto Seeding | If enabled, seed number for the random number generator is automatically determined |
| Random Number Seed | Seed number for the random number generator to generate the macroparticles |
| Integration Step | Longitudinal step normalized by the undulator period to solve the FEL equation. Also refer to Integration Step. |
| Steps/Segment | Steps in a single undulator segment |
| Steps in Drift Section | Steps in a single drift section |
| Total Beamlets | Total number of beamlets and number of macroparticles in a single beamlet. For details, refer to Structure of a Beamlet. |
| Particles/Beamlet | |
| Max. Beamlets/Slice | Maximum number of beamlets in a single slice |
| Beamlets/Slice | |
| Max. Harmonic | Maximum harmonic number to be considered |
| Spatial Window/σ | Specifies the spatial range to evaluate the radiation field |
| Grid Points Level | Specifies the number of spatial grid points |
| Grid Points | |
| Temporal Window (m) | Temporal window of the electron bunch |
| Bunch Position (m) | Slice position in the electron bunch |
| Total Slices | Total number of slices |
| Enable Parallel Computing | Enables the parallel computing by MPI. Note that MPI environment should be installed (Open MPI for LINUX and macOS, MS-MPI for MS-Windows) and path to mpiexec should be set. |
| Number of Processes | Number of MPI processes |
Simulation Mode
Specifies the simulation mode. Details are summarized below. Also refer to Simulation Modes
| Type | Detail |
| Steady State | The simulation is performed in a steady-state mode. |
| Time Dependent | The simulation is performed in a time-dependent mode. |
| Cyclic | As the above, but the radiation field slipped out of the head of the temporal window of the electron bunch is reused, namely shifted to the tail. This is convenient when doing a simulation in a limited temporal window of a long electron bunch. |
Simulation Option
Enables an option to be used in the simulation.
| Type | Detail |
| N.A. | No option is used. |
| Gaussian Mode | Apply a method to accelerate the numerical convergence in FEL simulations [12]. |
| Disable Quiet Loading | Turn off the quiet loading scheme to model the shot noize [13]. |
| Kill Shotnoize | Artifically disable the shot noize effect; the macroparticles are longitudinally arranged with a regular interval and thus no radiation (including spontaneous one) is initially generated. |
| Real Electron Number | Use the real number of electrons, which are randomly distributed in the longitudinal direction. |
Integration Step
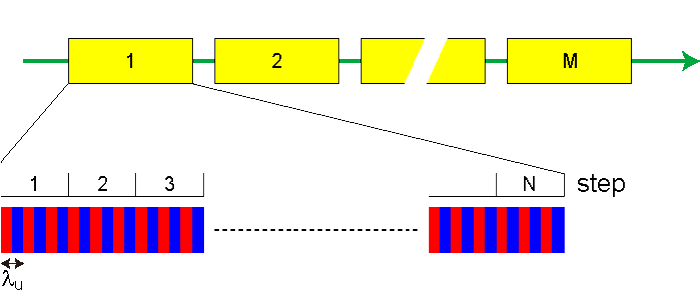
Structure of a Beamlet
In general, the number of macroparticles to be used in an FEL simulation is much less than the real number of electrons in the electron beam. This brings a problem of artificial (false) microbunching if the macroparticles are distributed randomly in the phase space. In order to solve the problem without increasing the number of macroparticles, Fawley's algorithm [13] of initial beam loading is implemented, in which all the macroparticles are divided into "beamlets" with the longitudinal length equal to the lasing wavelength; the macroparticles in the same beamlet have the identical initial position (x,y), angle (x',y') and energy (γ), and distributes almost evenly along the longitudinal axis with some perturbation to represent the shot noise.
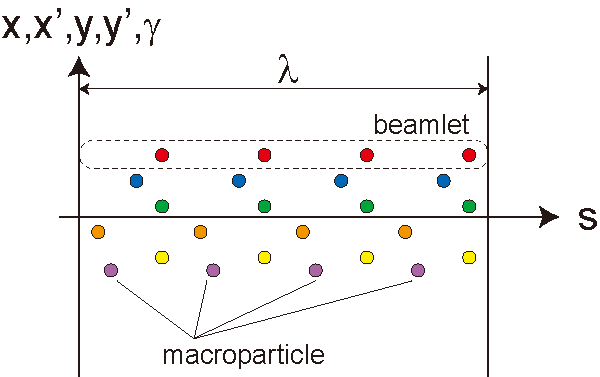
The motion of a specific macroparticle traveling along the undulator line is given by solving the equation of motion. The longitudinal position s and energy γ of each macroparticle are given by solving a pair of equations describing the interaction with radiation and undulator field.
In contrast, the transverse coordinate (x,y,x',y') of each macroparticle is determined by a linear transfer matrix along the undulator line. For example, the horizontal coordinate at the n-th step is given by
\[ \left(\begin{array}{c}x_n \\ x'_n \\ 1 \\ \end{array} \right)=\left(\begin{array}{ccc}C_n & S_n & D_n \\ C'_n & S'_n & D'_n \\ 0 & 0 & 1 \\ \end{array} \right)\left(\begin{array}{c}x_0 \\ x'_0 \\ 1 \\ \end{array} \right), \]
where C, S, and D are linear matrix elements defined by the arrangement of the focusing magnets in the undulator line, and subscript 0 means the value at the undulator entrance. Note that effects due to the vaiation of γ along the undulator line, which comes from the interaction with radiation, are supposed to be negligibly small compared to other factors (emittance etc.). As a result, the macroparticles in the same beamlet always have identical transverse coordinate (x,y,x',y'). As a reslt, we need (4+2Np)Nb variables to describe the motion of whole macroparticls in the 6-dimensional phase space; here Np is the number of macroparticls in a single beamlet specified by "Particles/Beamlet", while Nb is the number of beamlets to be used in the simulation specified by "Total Beamlets".
Data Dump Configurations
The parameters and configurations in the "Data Dump Configurations" subpanel are related to the conditions to export the simulation results. Detals are summarized below.
| Parameter/Option | Detail |
| Temporal Profile | Specifies if the related radiation profiles are saved in the output JSON file. |
| Spectral Profile | |
| Spatial Profile | |
| Angular Profile | |
| Output Interval | Step interval to evaluate the above profiles. Note the results at the final step are always exported. For example, data export is done at the steps of the 1st, 4th, ... 97th and 100th, if the total number of steps is 100 and this parameter is set at 3. |
| Particle Data | Saves the macroparticle raw data in a binary file (*.par). |
| Radiation Data | Saves the radiation raw data in a binary file (*.fld). |
| Output Steps | Specifies the steps to save the raw data. For details, refer to Steps to Export the Raw Data. |
| Initial Segment | Initial segment and segment interval to save the raw data. |
| Segment Step | |
| Step Interval | Step interval to save the raw data, if "Regular Interval" is chosen for "Output Steps". Note the results at the final step are always exported as explained Output Interval. In practice, too fine step interval is not recommended, because the file size to store the raw data can be huge. |
| Particle Data Size (MB) | Approximate file size of the birary file to save the raw data. |
| Radiation Data Size (MB) |
Steps to Export the Raw Data
Options to specify the steps to export the raw data are summarized below.
| Type | Details |
| All Segments | Export at the exit of every undulator segments. |
| Specific Segments | Export at the exit of specified undulator segments. |
| Final Step | Export once at the exit of the undulator line. |
| Regular Interval | Export with a regular step interval. Note that data at the final step is always exported. |
Output File
The parameters and configurations in the "Output File" subpanel are related to the name of the output JSON file and binary files to store the raw data. Detals are summarized below.
| Parameter/Option | Detail |
| Folder | Input the path to the output JSON file in [Folder], a prefix text in [Prefix], and a serial number in [Serial Number]. The data name is given as [Folder]/[Prefix]-[Serial Number], like "/Users/data/test-1", where "/Users/data", "test", and 1 refer to [Folder], [Prefix] and [Serial Number]. Note that the serial number can be -1 (negative), in which case it is not attached to the data name. |
| Prefix | |
| Serial Number |
FEL Performance
The performances of the FEL system evaluated using analytical and empirical expressions are shown in the "FEL Performance" subpanel. Detals are summarized below.
| Parameter/Option | Detail |
| Shotnoise Power (W) | Power of the shot noise to launch the SASE amplification process |
| ρ | Pierce parameter; refer to [2] for details |
| Lg1D,3D (m) | Gain length evaluated with an analytical formula under 1D approximation (1D), and with a more rigorous expression to consider adverse effects (3D)[4] for details |
| Psat (GW) | Satulation power and undulator length to reach saturation |
| Lsat (m) | |
| Pulse Energy (J) | Pulse energy at the saturation |
| ℏω1st (eV) | Fundamental photon energy and wavelength |
| λ1st (nm) | |
| FWHM Bandwidth | Bandwidth of the FEL radiation; refer to [3] for details. |
| Σx,y (mm) | RMS source size and angular divergence of the FEL radiation |
| Σx',y' (mrad) |
Pre-Processing
The "Pre-Processing" tab panel assists the pre-processing, i.e., visualization and configuration of the simulation conditions, which are not displayed in the subpanels explained above. Six pre-processing operations are available: "Graphical Plot", "Import Custom Data", "Analyze Particle Data", "Simplified Microbunch Evaluation", "Arrange Undulator Data" and "Optimize Lattice Function". Detail of each operation is explained below.
Graphical Plot
A number of items can be visualized to verify if the parameters and configurations specified in the subpanels are correct.
Items that can be visualized are listed in the left; just click one of them for visualization. Note that several of them are available only under specific conditions, which are summarized below. Note that "Export as ASCII" exports the current data set and saves as an ASCII file (select a file name in the dialog box), while "Duplicate Plot" creates a new window to duplicate the plot.
| Name in Pre-Processing Subpanel | Details | Available Condition | |
| Undulator | Field Distribution | Magnetic field distribution along the longitudinal axis in a specific segment. | |
| 1st Integral | 1st integrals of a specific segment, corresponding to the transverse velocity of an electron moving in the undulator | ||
| 2nd Integral | 2nd integrals of a specific segment, corresponding to the electron trajectory | ||
| Phase Error | Phase error [14] evaluated as a function the magnet pole number. Note that the number of end poles (used for the orbit adjustment and should be eliminated for the phase error evaluation) is automatically determined; to be specific, those with the peak field less than 95% of the average are ignored. | "Undulator Model" is not Ideal | |
| K Value Trend | Variation of the K value along the undulator line | "Tapering" is not N.A. | |
| Lattice | betatron Functions | betatron functions along the undulator line | |
| Optimization | Optimization of the focusing strength and initial Twiss parameters | ||
| Focusing Strength | Focusing strength along the undulator line | ||
| Wakefield | Wakefield Temporal Profile | Wakefield along the electron bunch | "Wakefield On" is true |
| Energy Distribution | Variation of the energy distribution of the electron beam | "Wakefield On" is true | |
| Others | Dispersion | Dispersion functions induced by injection and kick errors | "BPM Alignment" is not Ideal, "e- Injection Error" is true, or "Single Kick" is true |
| Mono. Spectrum | Complex reflectance of the monochromator | "Monochromator Type" is not N.A. | |
Import Custom Data
Under a specific simulation condition, the user should prepare custom data and import it. For example, a data file containing the current profile should be imported, if "Current Profile" is chosen for "Bunch Profile" option.
Details of importing the data set are summarized below. The data file to be imported should be an ASCII file, where "Values" are given as a function of "Independent Variable". "Condition" defines the condition in which the relevant "Data Type" should be imported. To import the data file, select "Item to Visualize" in the list, click Import button, and select the data file, or drag-and-drop the file in the area for visualization (grey-painted region in the above example).
| Data Type | Independent Variable | Values | Condition | Item to Visualize |
| Slice parameters (current, electron energy, emittance, etc.) | s | I , Energy , Energy Spread, εx , εy , βx , βy , αx, αy, <x> , <y> , <x'> , <y'> | "Bunch Profile" = "Slice Parameters" | Current Profile Slice Emittance ... |
| Current profile | s | I | "Bunch Profile" = "Current Profile" | Current Profile |
| Electron density in the (E-t) phase space | s , Δγ/γ | j | "Bunch Profile" = "E-t Profile" | E-t Distribution |
| Wakefield function by a point unit charge | s | Point-Charge Wake | "Additional Custom Wake" = "true" | Custom Wakefield |
| Complex reflectance of the monochromator | Photon Energy | Transmission Real, Transmission Imaginary | "Monochromator Type" = "Customize" | Custom Monochromator |
Meanings of "Values" and "Independent Variable" are as follows with the units to be used in the data file. Note that the units of "s" and "Energy" should be chosen before importing the data file, in a dialog box to popup by clicking "Edit Units" button.
- s: longitudinal position along the electron bunch
- Energy: slice electron energy
- Energy Spread: RMS slice energy spread (dimensionless)
- I: beam current (A)
- εx,y: normalized emittance (mm.mrad)
- βx,y: β Twiss parameter (m)
- αx,y: α Twiss parameter (dimensionless)
- <x,y>: Offsset position (m)
- <x',y'>: Offset angle (rad)
- Δγ/γ: normalized energy deviation (dimensionless)
- j: beam current density (A/100%)
The unit of j may need to be explained; it is given as the current per unit energy band; in a mathematical form, \[I(t)=\int j\left(t, \frac{\Delta\gamma}{\gamma}\right) d\frac{\Delta\gamma}{\gamma},\] and thus is given in the unit of Ampere in 100% energy band.
The data file should be an ASCII file with the format as follows. For the 1-dimensional (1D) data (current profile as an example),
s Current
-4.00000e-2 1.61861e+2
-3.99973e-2 1.61904e+2
-3.99947e-2 1.61947e+2
-3.99920e-2 1.61990e+2
(omitted)
3.99919e-2 1.61992e+2
3.99946e-2 1.61949e+2
3.99972e-2 1.61905e+2
3.99999e-2 1.61862e+2
where the 1st line (title) is optional. In the above format, the interval of the independent variable (s) does not have to be necessarily constant, which is not the case for the 2D data; the format should as follows
time DE/E j
-1.0e-3 -0.01 0.001
-0.9e-3 -0.01 0.002
-0.8e-3 -0.01 0.003
(omitted)
0.8e-3 -0.01 0.003
0.9e-3 -0.01 0.002
1.0e-3 -0.01 0.001
-1.0e-3 -0.008 0.001
-0.9e-3 -0.008 0.002
-0.8e-3 -0.008 0.003
(omitted)
0.8e-3 -0.008 0.003
0.9e-3 -0.008 0.002
1.0e-3 -0.008 0.001
(omitted)
-1.0e-3 0.01 0.001
-0.9e-3 0.01 0.002
-0.8e-3 0.01 0.003
(omitted)
0.8e-3 0.01 0.003
0.9e-3 0.01 0.002
1.0e-3 0.01 0.001
For reference, such a data format is created in the C/C++ language as follows.
for(n = 0; n < N; n++){
for(m = 0; m < M; m++){
cout << t[m] << " " << de[n] << " " << j[m][n] << endl;
}
}Note that the order of the "for loop" is arbitrary; the 1st and 2nd lines can be swapped in the above example.
After preparing the ASCII file, click "Import" button and specify the file name in the dialog box to import it. The unit of each item should be chosen before importing, in the Edit Units dialog box that pops up by running command. Note that the unit of the imported data cannot be changed, so you need to import the data again with the correct unit in case a wrong unit has been chosen.
Analyze Particle Data
"Analyze Particle Data" pre-processing operation is available when "Particle Distribution" is selected as "Bunch Profile", as shown below. Note that the user needs to specify in advance the data file containing the macroparticle positions in the 6D phase space, which is usually generated by another simulation code. Refer to Particle Distribution for details.
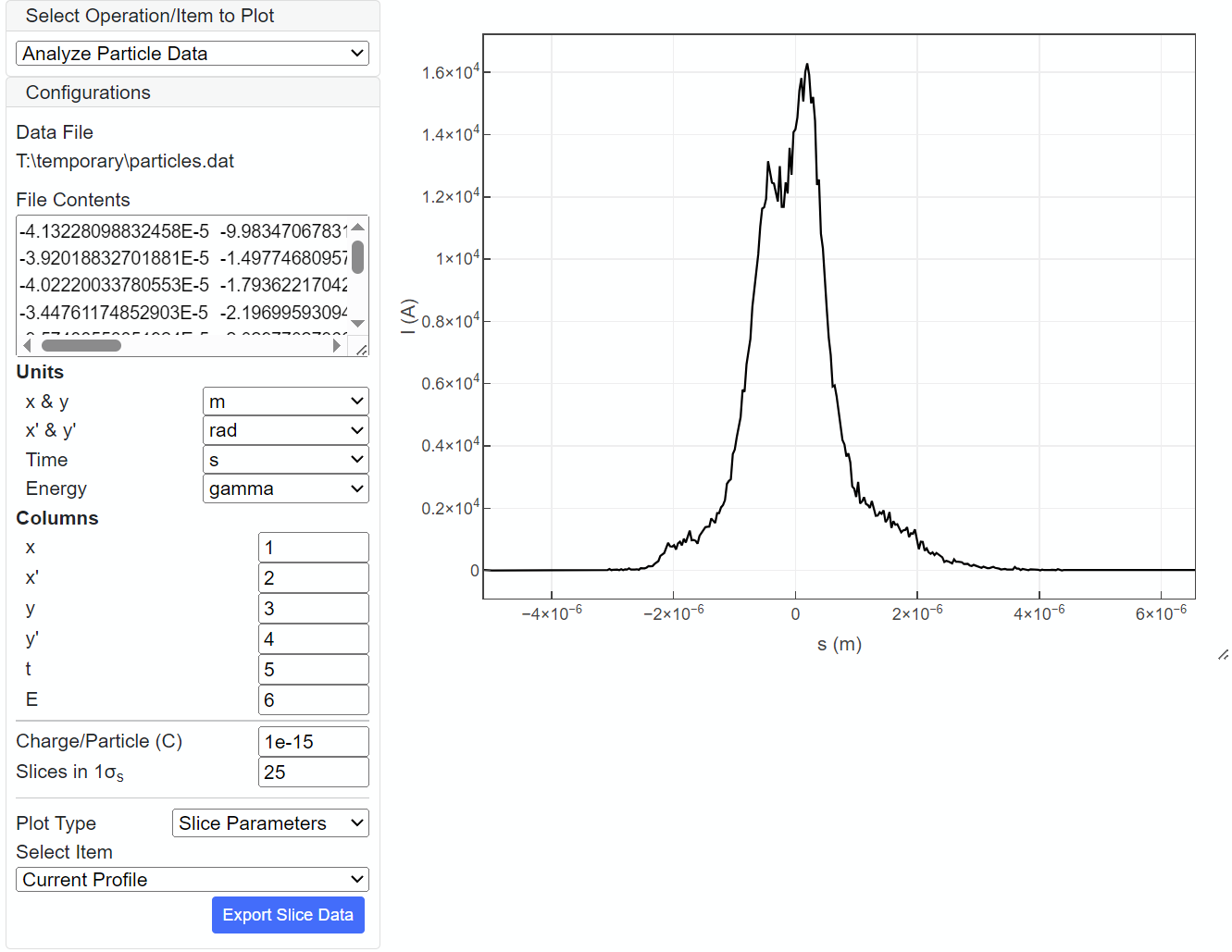
Once "Particle Distribution" is specified, SIMPLEX automatically loads the file to analyze the particle data. For convenience, part of the data file is shown in "File Contents" Select the unit and column index for each coordinate variable of the 6D (x,x',y,y',t,E) phase space and input relevant parameters. In the above example, each macroparticle has the charge of 3.5fC with "x" coordinate (horizontal position) located in the 1st column and the unit of m. Note that"Slices in 1σs" specifies the number of bins in the RMS bunch length, to be used for data analysis. Then, additional configurations appear in the GUI to specify how to visualize the results of analysis. In the above example, the current profile is plotted. Upon revision of the configurations above, SIMPLEX automatically analyzes the data with the new configuration and visualizes the result.
Besides the slice parameters shown in the above example, macroparticle distributions can be directly plotted. For example, distribution in the (E-t) phase space is plotted in the figure below.
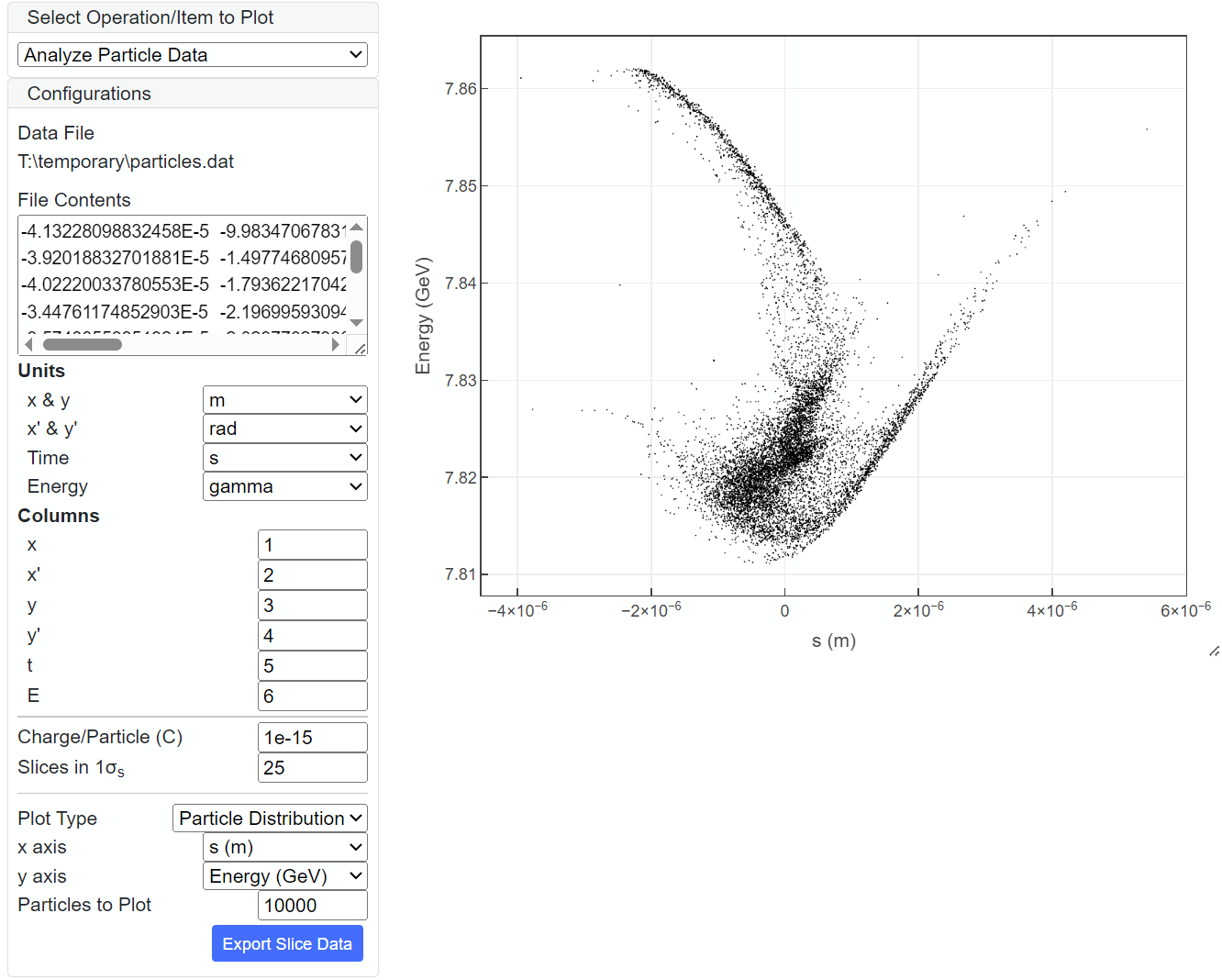
The result of analysis can be exported and saved as an ASCII file; "Export Selected" exports the data currently plotted, while "Export Slice Data" exports the whole slice data (slice paramters vs. s). The exported data file can be used later as the custom data file for slice parameters or current profile.
Simplified Microbunch Evaluation
"Simplified Microbunch Evaluation" pre-processing operation is available for seeded FEL simulations, namely, when options other than "N.A." is chosen as "Seed Light". This is to roughly evaluate the energy modulation in the electron beam through interaction with the seed light. Unlike the rigorous FEL simulation, it neglects the amplification of radiation and other 3D effects coming from the finite emittance of the electron beam, and thus is much faster. Although the result may be less reliable, it can be used to roughly estimate the microbunching in the seeded FELs.
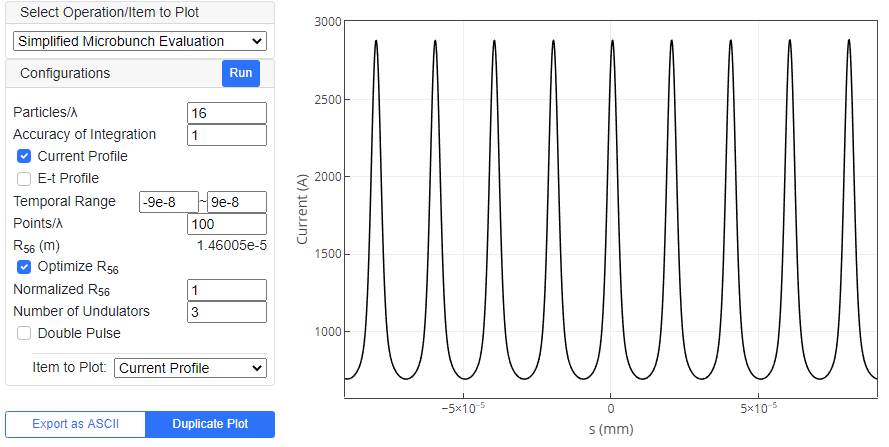
To start evaluating the microbunching, edit the parameters and configurations listed in the left side, and click "Run" button.
| Parameter/Option | Detail |
| Particles/λ | Number of macroparticles per fundamental wavelength |
| Accuracy of Integration | Accuracy of integration to evaluate the current profile. Given by a positive integer; the larger the more accurate. |
| Current Profile | Evaluate the current profile |
| E-t Profile | Evaluate the density in the (E,t) phase space |
| Temporal Range | Temporal range to evaluate the current and E-t profiles |
| Points/λ | Number of points per fundamental wavelength to evaluate the current and E-t profiles |
| Energy Range | Energy range to evaluate the E-t profile |
| Energy Points | Number of energy points to evaluate the E-t profile |
| R56 (m) | Longitudinal dispersion of a virtual chicane to be positioned after the undulator |
| Optimize R56 | If enabled, the longitudinal dispersion is given by R56=r×r56 where r56 is the dispersion to maximize the density modulation (peak current) to be automatically determined, while r is the normalized value that can be adjusted by the user. |
| Normalized R56 | |
| Number of Undulators | Number of undulator segments to be used for the numerical evaluation |
| Double Pulse | If enabled, another seed pulse with similar properties is supposed |
| Δλ/λ | Relative wavelength of the 2nd seed pulse |
| CEP (°) | Carrier envelope phase of the 2nd seed pulse |
| GDD (fs2) | Groupd delay dispersion of the 2nd seed pulse |
| TOD (fs2) | Third order dispersion of the 2nd seed pulse |
| Time Delay (fs) | Relative timing of the the 2nd seed pulse |
The results of evaluation can be plotted in three different ways (depending on the configurations): distribution of the reference particle in the (E-t) space, current profile, or E-t profile. Select one of the available plots.
Arrange Undulator Data
"Arrange Undulator Data" pre-processing operation is available when "Import Data" is selected as "Undulator Model". Example of the tab panel is shown below.
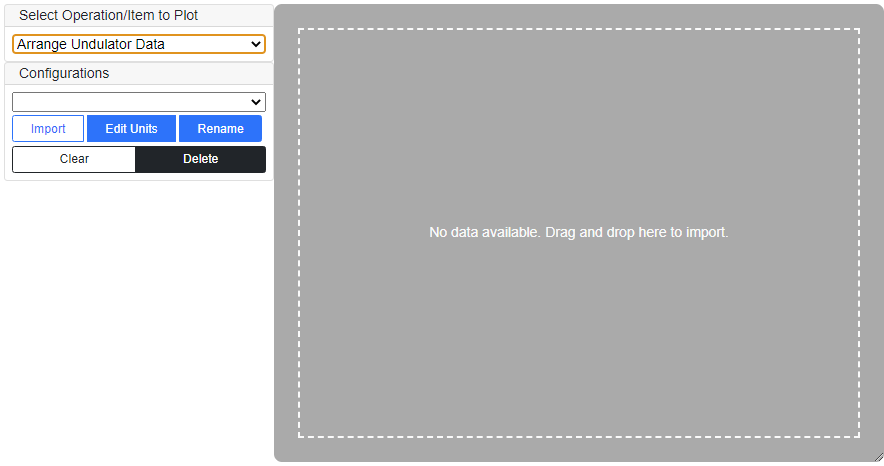
Click "Import" button and select the undulator data file or drag-and-drop the file in the area for visualization (grey-painted region in the above example). Be sure that the correct units are selected for the longitidunal position and magnetic field strength.
The imported data name can be changed by clicking "Rename" button and input a desired name in the modal dialog box. If a certain data set is not necessary any more, click "Delete" button to delete it. "Clear" button deletes all the imported data sets.
Optimize Lattice Function
"Optimize Lattice Function" pre-processing operation tries to optimize the betatron function through the undulator line.
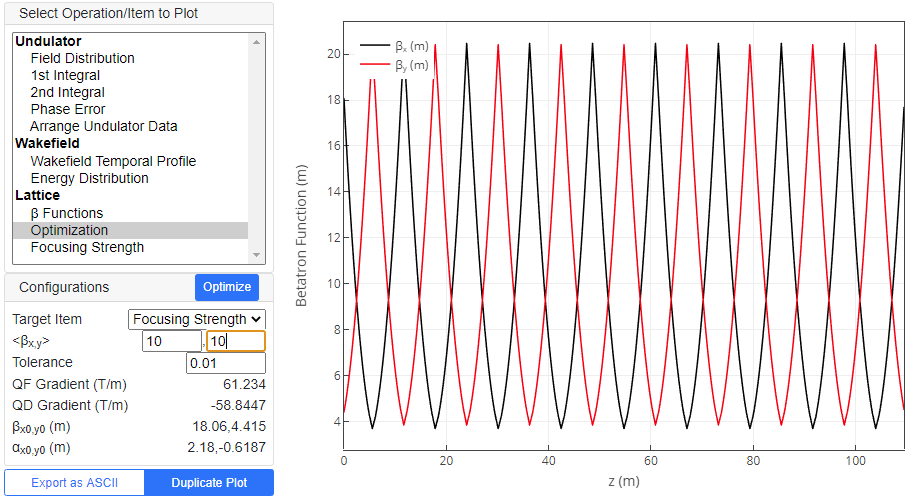
We have two points to take care in optimization: betatron matching and average over the whole undulator line. The former is related to the initial Twiss parameters, while the latter is determined by the strength of the quadrupole magnets. Click "Optimize" button to start the optimization process. After its completion, the betatron functions through the undulator line are plotted. Parameters needed for optimization are summarized below.
| Parameter/Option | Detail |
| Target Item | Target of optimization. "Average" optimizes both of the quadrupole strength and initial Twiss parameters, while "Matching" does the Twiss parameters alone, without changing the qudrupole strength from the current value. |
| <βx,y> | Target average betatron functions. After the optimization process, they are updated by the actual values given by the quadrupole strengths and initial Twiss parameters determined by the optimization, and thus are not necessarily the same as the input values. |
| Tolerance | Optimization tolerance of the average betatron function |
| QF Gradient (T/m) | Quadrupole strengths and initial Twiss parameters determined by the optimization process. Note that these values, which are shown upon completion of the optimization process, are automatically copied to the same parameters in Lattice parameter list. |
| QD Gradient (T/m) | |
| βx0,y0 (m) | |
| αx0,y0 (m) |
Note that the achieved values (<βx,y>) can be far from the target ones, in which case the structure of the undulator line (segment interval, lattice type, etc.) may not be consistent with the electron energy. Another issue is that no solutions are found for the initial Twiss parameters to satisfy the betatron matching condition, in which case the betatron functions (and thus the electron beam size) can diverge. Although the former issue may be probably acceptable, the latter one should be solved before starting a simulation.
Post-Processing
The "Pre-Processing" tab panel assists the post-processing, i.e., visualization of the simulation results. Upon completion of a simulation, the output JSON file is automatically loaded, or alternatively, the existing output JSON files can be imported by clicking "Import" button and specify its path.
Two post-processing operations are available: "Raw Data Processing" and "Visualization". The former post-processes the raw data saved in the binary files to evaluate certain items and save the results in a file to be used for visualization, while the latter configures the graphical plot to visualize the simulation results, including those obtained by Raw Data Processing.
Visualization
To visualize the simulation results such as the gain curve and radiation profiles saved in the output JSON file, and the post-processed data generated by Raw Data Processing, select "Data Type" and "Items to Plot" from the list. For 1D plot, also select "x axis" to specify the item for the x axis of the plot. The details of Available Data Type are summarized below.
| Name | Detail | Plot Type |
| Gain Curve | Values integrated over the whole electron bunch, such as the pulse energy of radiation and bunch factor, evaluated as a function of the longitudinal position along the undulator line. | 1D |
| Temporal Profile | Radiation power integrated over the whole solid angle vs. the bunch, | 1D animation |
| Spectral Profile | Spectrum of radiation integrated over the whole solid angle. | 1D animation |
| Spatial Profile | Spatial profile of radiation integrated over the whole slice. | 2D animation |
| Angular Profile | Angular profile of radiation integrated over the whole slice. Note that the angular coordinate is normalized by the reciprocal of a harmonic order. Namely, it is given as h×&theta, where h is the harmonic order. | 2D animation |
| Processed [s/n]: * | Result of Raw Data Processing. "s/n" stands for the serial number specified by the user (optional), and * means the target item for post-processing. | Depends on the configurations of Raw Data Processing |
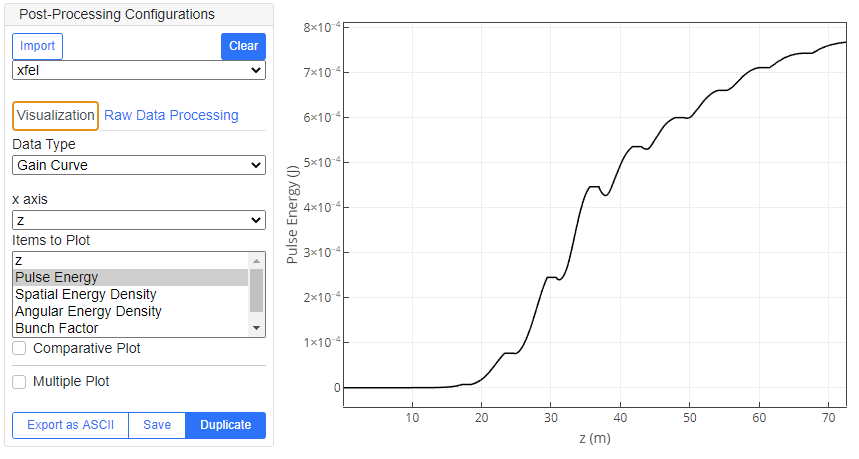
The functions of the three buttons in the bottom right are as follows."Export as ASCII" exports the current data set and saves as an ASCII file (select a file name in the dialog box), "Save" saves all the configurations of the plot as well as the data in a JSON file, which can be imported later by Load Post-Processed Result, and "Duplicate Plot" creates a new window to duplicate the plot.
The dimension of the plot depends on the selected data type and item. In a 1D plot, a desired area can be zoomed in by dragging. Other options are available to operate the plot, by clicking one of the small icons located in the right top of the plot. For details, refer to the documents about "Plotly" library to be found online. Besides the above options, the plot can be configured by clicking the "pencil" icon and edit the parameters in the modal dialog box to pop up. The details of the parameters are summarized below.
| Parameter/Option | Detail |
| Scale | Select the normalization scheme for animation plots. "For Each" means that the y(z) axis is normalized for each slide, while "By Maximum" means that it is normalized by a maximum value through the whole slides. |
| X-axis Scale | Select the linear/log scale for x and y axes. |
| Y-axis Scale | |
| Plot Type | Select the plot type for the 1D plot. |
| Symbol Size | Select the symbol size for the 1D plot. |
| Line Width | Select the line width for the 1D plot. |
| 2D Plot Type | Select the plot type for the 2D plot. |
| Color | Select the color of the surface plot. |
| Color Map | Select the color scale pattern of the surface plot. |
| Show Scale | Enable the color scale. |
| Wireframe | Add a wire frame on the surface plot. |
Comparative Plot
If more than one output JSON file is loaded, and/or Raw Data Processing has been performed before, "Comparative Plot" will be available depending on the simulation conditions. Select data names from the list to compare the results.
Multiple Plot
"Multiple Plot" creates more than one plot to view several results simultaneously. The difference from Comparative Plot is that this allows for selection of different items and variables, and the dimension of the plot can be different. Note, however, that the number of steps should be the same for an animation plot.
Plotting 3/4-Dimensional Data
Depending on the configurations and target items, Raw Data Processing pontentially generates 3D or 4D data, which cannot be plotted in a straightforward manner. In SIMPLEX, the 3D/4D data are "sliced" into a number of data sets and plotted as a 1D or 2D graph. Let f(z,s,x,y) be a function defined by 4 argument variables, z, s, x, and y. SIMPLEX offers several combinations for plotting and slicing variables. If a pair (x,y) is chosen as the plotting variable, then the data is sliced at each (z,s) position, and a 2D plot is created. Note that the coordinates of the slicing variables can be arbitrary chosen within the possible range, by dragging the sliders indicating their positions.
Raw Data Processing
General Instructions
To perform "Raw Data Processing", radiation/particle data should be saved, by enabling "Radiation Data"/"Particle Data" option before starting a simualtion. Then, as explained in Arrange the Output File, two types of binary files "*.fld" and "*.par" are generated when a simulation is completed. In the former file, complex amplitudes of radiation are saved at specified steps, which are then retrieved to evaluate three different items: "Radiation Power", "Photon Flux" and "Complex Amplitude". In the latter file, coordinate variables of all the macroparticles in the 6D phase space are saved, which are then retrieved to evaluate "Bunch Factor" and "Energy Distribution" , as well as "Particle Motion".
Parameters and configurations
| Parameter/Option | Detail |
| Target | Target item to be evaluate by Raw Data Processing |
| Real/Imaginary Part | Select real/imaginary component for an item given by a complex number |
| Ex,y | Select horizontal/vertical component for the radiation field |
| S1 (Horizontal) | If enabled, relevant Stoke parameters are evaluated |
| S2 (45-deg.) | |
| S3 (Circular) | |
| Harmonic Number | Target harmonic number |
| Zone | Select the observation zone (near or far field) |
| Coordinate System | Select the coordinates to retrieve the macroparticle data |
| Step | Select how to specify the range of interest (ROI) for respective variables. "Whole" means that the whole range used in the simulation is assumed, while "Set Window" put a window on the ROI. |
| Slice | |
| Photon Energy | |
| Space | |
| Angle | |
| Step (z) | Range of the step and corresponding range of the longitudinal coordinate (z) |
| z (m) | |
| Slice (s) | Range of the slice and corresponding range of the bunch position (s) |
| s (m) | |
| Energy (ℏω) | Range of the photon energy |
| ℏω (eV) | |
| Spatial (x,y) | Half range of the transverse position |
| Δx,y (mm) | |
| Angle | Half range of the observation angle |
| Δx',y' (mrad) | |
| Integration (s) | Enable integration over the ROI |
| Integration (x,y) | |
| Integration (x',y') | |
| R56 (m) | Virtual longitudinal dispersion to be added after each step. This is convenient to evaluate the reasonable R56 to enhance the microbunching. |
| Data Serial Number | Serial number to be attached to the data file to save the post-processed data. |
Perform Post-Processing
Once the configurations have been set, click "Run Data-Processing" to start post-processing. A progress bar appears to inform its progress. When completed, a new file is created to save the post-processed data, with the name "*-pp(s/n).json", wherer * is the data name and s/n is the serial number if specified (>0). This file is automatically loaded and the data is visualized. For details of how to visualize the data, refer to Visualization.
File Format
Besides the operation based on the GUI, SIMPLEX (more precisely, the solver) can be utilized to communicate with external codes for the so-called start-to-end simulations. This actually requires the knowledge of the format of the input and output files, which is explained in the followings.
JSON Format
To deal with the many parameters and options, SIMPLEX utilizes the JSON (JavaScript Object Notation) format, which is described by a number of "objects". The object contains a number of "pairs" formed by a "key" and "value", separated by a colon ":", and should be enclosed by a curly bracket {}. The value can be one of the followings: number, array, string and (another) object. An example of the SIMPLEX input file is as follows.
{
"Electron Beam": {
"Bunch Profile": "Gaussian",
"Electron Energy (GeV)": 1,
....
},
"Seed Light": {
"Seed Light": "Gaussian Beam",
"Peak Power (W)": 1000000,
....
},
"Undulator": {
"Undulator Type": "Linear",
"K Value": 2.03161,
....
},
"Lattice": {
"Lattice Type": "FUDU (QF-U-QD-U)",
"QF Gradient (T/m)": 5.89896,
....
},
....
}In this example, four JSON objects are found, whose keys are "Electron Beam", "Seed Light", "Undulator", and "Lattice". The value of each object is also an object, which actually specifies the parameters and options, such as "Energy (GeV)": 1, denoting the energy of the electron beam to be 1 GeV.
For details of the JSON format, please refer to any document available online or found in a text book.
Input Format
The input file to be loaded by the solver should have 4 JSON objects: "Electron Beam", "Seed Light", "Undulator", and"Simulation Conditions". Details of each object are summarized below, where "GUI Notation" is the parameter name displayed in the GUI panel, "Key" is the name of the key to be used in the input file, "Format" is the format of the value, and "Default" is the default value. Note that the key name can be either of the "Full" or "Simplified" expression.
Electron Beam Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Bunch Profile | Bunch Profile | bmprofile | Select from: "Gaussian" "Boxcar" "SIMPLEX Output" "[object Object]" |
Gaussian |
| Slice Parameters | Slice Parameters | slicefile | object | |
| Current Profile | Current Profile | currfile | object | |
| E-t Profile | E-t Profile | etfile | object | |
| Particle Distribution | Particle Distribution | partfile | file name | |
| Electron Energy (GeV) | Electron Energy (GeV) | eenergy | number | 8 |
| RMS Bunch Length (m) | RMS Bunch Length (m) | bunchleng | number | 0.000005 |
| Bunch Length (m) | Bunch Length (m) | bunchlenr | number | 0.00001 |
| Bunch Charge (nC) | Bunch Charge (nC) | bunchcharge | number | 0.15 |
| εx,y (mm.mrad) | ε<sub>x,y</sub> (mm.mrad) | emitt | array | [0.4, 0.4] |
| RMS Energy Spread | RMS Energy Spread | espread | number | 0.0001 |
| Energy Chirp (m-1) | Energy Chirp (m<sup>-1</sup>) | echirp | number | 0 |
| ηx,y (m) | η<sub>x,y</sub> (m) | eta | array | [0, 0] |
| Peak Current (A) | Peak Current (A) | pkcurr | number | 3588 |
| σ0x,0y (mm) | σ<sub>0x,0y</sub> (mm) | ebmsize | array | [0.02747649154431839, 0.023085244683238686] |
| σ0x',0y' (mrad) | σ<sub>0x',0y'</sub> (mrad) | ediv | array | [0.0013503330424008604, 0.0013881904776490983] |
| <σx,y> (mm) | <σ<sub>x,y</sub>> (mm) | avgsize | array | [, ] |
| R56 (m) | R<sub>56</sub> (m) | r56 | number | 0 |
| Bunch Twiss Prms. | Bunch Twiss Prms. | twissbunch | Select from: "Projected" "Sliced (Optimized)" "Sliced (Custom)" |
Projected |
| Slice Position (m) | Slice Position (m) | twisspos | number | 0 |
Seed Light Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Seed Light | Seed Light | seedprofile | Select from: "N.A." "Gaussian Beam" "Chirp Pulse" "SIMPLEX Output" |
N.A. |
| Peak Power (W) | Peak Power (W) | pkpower | number | 1000000000 |
| Δλ/λ1 | Δλ/λ<sub>1</sub> | relwavelen | number | 0 |
| FWHM Pulse Length (fs) | FWHM Pulse Length (fs) | pulselen | number | 40 |
| FWHM Spot Size (mm) | FWHM Spot Size (mm) | spotsize | number | 0.05 |
| Position of Waist (m) | Position of Waist (m) | waistpos | number | 0 |
| Relative Time (fs) | Relative Time (fs) | timing | number | 0 |
| Carrier Envelope Phase (°) | Carrier Envelope Phase (°) | CEP | number | 0 |
| GDD (fs2) | GDD (fs<sup>2</sup>) | gdd | number | 0 |
| TOD (fs3) | TOD (fs<sup>3</sup>) | tod | number | 0 |
| Phase Offset (°) | Phase Offset (°) | phase | number | 0 |
SIMPLEX Output Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Data Name | Data Name | spxfile | file name | |
| Step Index to Retrieve Data | Step Index to Retrieve Data | spxstep | integer | 0 |
| Exported Positions (m) | Exported Positions (m) | spxstepzarr | array | [] |
| Total Beamlets | Total Beamlets | bmletsout | number | 0 |
| Particles/Beamlet | Particles/Beamlet | paticlesout | number | 0 |
| Drift Length (m) | Drift Length (m) | matching | number | 1 |
| Photon Energy (eV) | Photon Energy (eV) | spxenergy | number | 10 |
Undulator Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Undulator Type | Undulator Type | utype | Select from: "Linear" "Helical" "Elliptical" "Multi-Harmonic" |
Linear |
| K Value | K Value | K | number | 2.18 |
| K⊥ | K<sub>⊥</sub> | Kperp | number | 2.18 |
| Multi-Harmonic Contents | Multi-Harmonic Contents | multiharm | array | [] |
| tan-1(Kx/Ky) (°) | tan<sup>-1</sup>(K<sub>x</sub>/K<sub>y</sub>) (°) | epukratio | number | 30 |
| λu (mm) | λ<sub>u</sub> (mm) | lu | number | 18 |
| Length/Segment (m) | Length/Segment (m) | length | number | 5 |
| Number of Segments | Number of Segments | segments | integer | 18 |
| Segment Interval | Segment Interval | interval | number | 6.15 |
| Extra Slippage (°) | Extra Slippage (°) | exslippage | number | 0 |
| Tapering | Tapering | taper | Select from: "N.A." "Stair-Like" "Continuous" "Custom" |
N.A. |
| Optimization | Optimization | opttype | Select from: "N.A." "Compensate Wake" "Projection" "Slice" |
N.A. |
| Target Slice Position (m) | Target Slice Position (m) | slicepos | number | 0 |
| Initial Segment | Initial Segment | initial | integer | 10 |
| Increment Segment Interval | Increment Segment Interval | incrseg | integer | 2 |
| Base Linear Taper (m-1) | Base Linear Taper (m<sup>-1</sup>) | base | number | 0.001 |
| Taper Increment (m-1) | Taper Increment (m<sup>-1</sup>) | incrtaper | number | 0.001 |
| K Value@Exit | K Value@Exit | Kexit | number | 2.024 |
| Resonance Detuning@Exit | Resonance Detuning@Exit | detune | number | 0.08 |
| Custom Taper | Custom Taper | tapercustom | array | [] |
| Undulator Model | Undulator Model | umodel | Select from: "Ideal" "Specify Error" "Import Data" |
Ideal |
| Random Number Auto Seeding | Random Number Auto Seeding | umautoseed | boolean | false |
| Random Number Seed | Random Number Seed | umrandseed | integer | 1 |
| σφ (°) | σ<sub>φ</sub> (°) | phaseerr | number | 5 |
| σB/B (%) | σ<sub>B</sub>/B (%) | berr | number | 0.5 |
| σx,y (mm) | σ<sub>x,y</sub> (mm) | xyerr | array | [0.001, 0.001] |
| Apply to All Segments | Apply to All Segments | allsegment | boolean | false |
| Target Segment | Target Segment | tgtsegment | integer | 1 |
Lattice Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Lattice Type | Lattice Type | ltype | Select from: "FUDU (QF-U-QD-U)" "DUFU (QD-U-QF-U)" "FUFU (QF-U-QF-U)" "Doublet (QF-QD-U)" "Triplet (QF-QD-QF-U)" "Undulator Combined" |
FUDU (QF-U-QD-U) |
| QF Gradient (T/m) | QF Gradient (T/m) | qfg | number | 18.99 |
| QD Gradient (T/m) | QD Gradient (T/m) | qdg | number | -17.44 |
| QF Length (m) | QF Length (m) | qfl | number | 0.1 |
| QD Length (m) | QD Length (m) | qdl | number | 0.1 |
| QF-QD Distance (m) | QF-QD Distance (m) | dist | number | 0.02 |
| # Periods/Segment | # Periods/Segment | lperiods | integer | 18 |
| βx0,y0 (m) | β<sub>x0,y0</sub> (m) | betaxy0 | array | [29.5483, 20.8583] |
| αx0,y0 (m) | α<sub>x0,y0</sub> (m) | alphaxy0 | array | [1.05297, -0.757107] |
Wakefield Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Wakefield On | Wakefield On | wakeon | boolean | false |
| Aperture (m) | Aperture (m) | aperture | number | 0.0035 |
| Resistive | Resistive | resistive | boolean | false |
| Resistivity (Ωm) | Resistivity (Ωm) | resistivity | number | 1.68e-8 |
| Relaxation Time (sec) | Relaxation Time (sec) | relaxtime | number | 8e-15 |
| Parallel Plate | Parallel Plate | paralell | boolean | true |
| Surface Roughness | Surface Roughness | roughness | boolean | false |
| RMS Height (m) | RMS Height (m) | height | number | 0.000002 |
| Correlation Length (m) | Correlation Length (m) | corrlen | number | 0.00004 |
| Dielectric Layer | Dielectric Layer | dielec | boolean | false |
| ε/ε0 | ε/ε<sub>0</sub> | permit | number | 2 |
| Thickness (m) | Thickness (m) | thickness | number | 2e-8 |
| Space Charge | Space Charge | spcharge | boolean | false |
| Additional Custom Wake | Additional Custom Wake | wakecustom | boolean | false |
| Wakefield Data | Wakefield Data | wakecustomdata | object | |
Alignment Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Undulator Alignment | Undulator Alignment | ualign | Select from: "Ideal" "Specify Tolerance" "Specify Offset" |
Ideal |
| ΔK Tolerance | ΔK Tolerance | Ktol | number | 0 |
| Slippage Tolerance (°) | Slippage Tolerance (°) | sliptol | number | 0 |
| Offset for Each Segment | Offset for Each Segment | sigsegment | array | [] |
| BPM Alignment | BPM Alignment | BPMalign | Select from: "Ideal" "Specify Tolerance" |
Ideal |
| x,y Tolerance (mm) | x,y Tolerance (mm) | xytol | array | [0.05, 0.05] |
| Random Number Auto Seeding | Random Number Auto Seeding | alautoseed | boolean | false |
| Random Number Seed | Random Number Seed | alrandseed | integer | 1 |
Chicane Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Chicane On | Chicane On | chicaneon | boolean | false |
| Electron Delay (fs) | Electron Delay (fs) | delay | number | 10 |
| Chicane Position | Chicane Position | chpos | integer | 10 |
| Rearrange After Chicane | Rearrange After Chicane | rearrange | boolean | false |
| Monochromator Type | Monochromator Type | monotype | Select from: "N.A." "Transmission" "Reflection" "Customize" |
N.A. |
| Monochromator Data | Monochromator Data | monodata | object | |
| Crystal Type | Crystal Type | xtaltype | Select from: "Diamond (400)" "Diamond (220)" "Silicon (111)" "Customize" |
Diamond (400) |
| ℏω (eV) | ℏω (eV) | monoenergy | number | 10000 |
| |Fg| | |F<sub>g</sub>| | formfactor | number | 12.72 |
| Lattice Spacing (nm) | Lattice Spacing (nm) | latticespace | number | 0.089175 |
| Unit Cell Volume (nm3) | Unit Cell Volume (nm<sup>3</sup>) | unitvol | number | 0.045385 |
| Crystal Thickness (mm) | Crystal Thickness (mm) | xtalthickness | number | 0.1 |
| Relative Timing (fs) | Relative Timing (fs) | reltiming | number | 0 |
Dispersion Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| e- Injection Error | e<sup>-</sup> Injection Error | einjec | boolean | false |
| Δx,y (mm) | Δx,y (mm) | exy | array | [0, 0] |
| Δx',y' (mrad) | Δx',y' (mrad) | exyp | array | [0, 0] |
| Single Kick | Single Kick | kick | boolean | false |
| Kick Position (m) | Kick Position (m) | kickpos | number | 0 |
| Kick Angle x,y (mrad) | Kick Angle x,y (mrad) | kickangle | array | [0, 0] |
| Seed Injection Error | Seed Injection Error | sinjec | boolean | false |
| Seed Δx,y (mm) | Seed Δx,y (mm) | sxy | array | [0, 0] |
| Seed Δx',y' (mrad) | Seed Δx',y' (mrad) | sxyp | array | [0, 0] |
Simulation Conditions Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Simulation Mode | Simulation Mode | simmode | Select from: "Time Dependent" "Steady State" "Cyclic" |
Time Dependent |
| Simulation Option | Simulation Option | simoption | Select from: "N.A." "Gaussian Mode" "Disable Quiet Loading" "Kill Shotnoize" "Real Electron Number" |
N.A. |
| Skip Wavefront Transfer Process | Skip Wavefront Transfer Process | skipwave | boolean | false |
| Auto Integration Step | Auto Integration Step | autostep | boolean | true |
| Random Number Auto Seeding | Random Number Auto Seeding | autoseed | boolean | false |
| Random Number Seed | Random Number Seed | randseed | integer | 1 |
| Integration Step | Integration Step | step | integer | 5 |
| Total Beamlets | Total Beamlets | beamlets | number | 10000000 |
| Total Electrons | Total Electrons | electrons | number | 10000000 |
| Max. Electrons/Slice | Max. Electrons/Slice | sliceels | number | 2000 |
| Electrons/Slice | Electrons/Slice | sliceelsss | number | 2000 |
| Max. Harmonic | Max. Harmonic | maxharmonic | integer | 1 |
| Particles/Beamlet | Particles/Beamlet | particles | integer | 4 |
| Spatial Window/σ | Spatial Window/σ | spatwin | integer | 4 |
| Temporal Window (m) | Temporal Window (m) | simrange | array | [-0.000006, 0.000006] |
| Bunch Position (m) | Bunch Position (m) | simpos | number | 0 |
| Enable Parallel Computing | Enable Parallel Computing | enablempi | boolean | false |
| Number of Processes | Number of Processes | mpiprocs | integer | 4 |
Data Dump Configurations Object
| GUI Notation | Key | Format | Default | |
| Full | Simplified | |||
| Radiation Profile Data | Radiation Profile Data | procdata | string | |
| Temporal Profile | Temporal Profile | temporal | boolean | false |
| Spectral Profile | Spectral Profile | spectral | boolean | false |
| Spatial Profile | Spatial Profile | spatial | boolean | false |
| Angular Profile | Angular Profile | angular | boolean | false |
| Output Interval | Output Interval | profstep | integer | 1 |
| Raw Data | Raw Data | rawdata | string | |
| Particle Data | Particle Data | particle | boolean | false |
| Radiation Data | Radiation Data | radiation | boolean | false |
| Output Steps | Output Steps | expstep | Select from: "All Segments" "Specific Segments" "Final Step" "Regular Interval" |
All Segments |
| Initial Segment | Initial Segment | iniseg | integer | 1 |
| Segment Step | Segment Step | segint | integer | 1 |
| Step Interval | Step Interval | stepinterv | integer | 1 |
Output Format
To facilitate further processing of the simulation result with other external codes, the structure of the output JSON file is explained below. Note that the order index (for example of an array, column, etc.) in the followings is defined as starting from "0", but not from "1".
"Input" Object
All the parameters and options are stored in this object with the same format as the Input Format. If the output JSON file is opened in the GUI (as an input parameter file), these parameters are displayed and can be used again.
Objects to Save the Simulation Results
The simulation results are saved in a number of objects with the keys of "Gain Curve", "Temporal Profile", "Spectral Profile", "Spatial Profile" and "Angular Profile" (last 4 are optional), and can be visualized by Visualization as mentioned in Post-Processing.
| Key | Details | Format |
| dimension | Dimension of the data | number |
| variables | Number of independent variables for each sliced data. If "dimension" is 3 and "variables" is 2, then the 2D data (for example, spatial profile of radiation) was generated repeatedly at each step. These conditions are used later for visualization | number |
| titles | Titles of individual arrays included in the "data" object. | array (1D) |
| units | Units of individual arrays included in the "data" object. | array (1D) |
| data | Main body of the simulation result data. | array (2D or 3D) |
The format of the "data" object (2D array) is as follows.
- 0th ~ (n-1)-th array: independent variables, where n is the dimension
- n-th ~ (n+m-1)-th array: simulation results, where m is the number of items. The length of each array corresponds to the product of the lengths of the independent variable arrays.
As an example, let us consider the "Gain Curve" object as follows
"Gain Curve": {
"dimension": 1,
"titles": ["z","Pulse Energy","Spatial Energy Density","Angular Energy Density","Bunch Factor","Energy Loss"],
"units": ["m","J","J/mm2","J/mrad2","-","J"],
"data": [
[0.18,0.36,...,72.33,72.51],
[3.78429e-09,7.57999e-09,...,0.000767857,0.000768138],
[2.75008e-06,5.28602e-06,...,0.23161,0.230191,0.229851],
[2.20925e-06,4.46296e-06,...,126.883,126.987,127.091],
[0.00195631,0.00195761,...,0.0183189,0.0178893],
[-5.80174e-09,-5.30405e-09,...,-0.000779005,-0.000777961]
]
},The data is composed of 1 independent variable (z) and 5 items (Pulse Energy etc.). The 0th array ([0.18,...]) corresponds to the longitudinal coordinate, and the 1st ([3.78429e-09,...]) to the pulse energy, etc.
In case the dimension is larger than 1 and thus more than one independent variables exist, the order index j of the item array is given as \[j=j_0+j_1N_0+j_2N_0N_1+\cdots,\] where $j_i$ and $N_i$ refer to the order index and number of data points of the $i$-th variable.
Binary File Format
Besides the output JSON file described above to summarize the simulation results, SIMPLEX optionally exports "raw data", namely, the distribution of macroparticles and complex amplitude of radiation. Because these data can be potentially large, they are saved in a binary format to save the required memory size. Besides being used as input data for another simulation with SIMPLEX, they can be retrieved as numerical data for other applications. For this purpose, the format of these binary files are described below. Note that all of the raw data are saved as float (4-byte) numbers instead of double (8-byte) ones to reduce the file size.
Configurations
To retrieve the binary data, its configurations are needed. In SIMPLEX, they are saved separately in the output file (*.json) as an object defined by "Raw Data Export" as follows.
{
"Raw Data Export": {
...,
"Steps (m)": [4.86,11.01,17.16,23.31,29.46,35.61],
"Slices (m)": [-9.99382e-07,-9.97437e-07,...,9.99382e-07],
"Grid Intervals (m,rad)": [2.13045e-06,1.23744e-06,4.75329e-07,8.18358e-07],
"Grid Points": [128,128],
"Beamlets": 500000,
"Particles/Beamlet": 4,
....
},
....
}In this example, the raw data are exported at 6 ($=K$) longitudinal positions defined as $z$ = 4.86, 11.01,... . The number of beamlets (=$N_b$) and particles per beamlet ($=N_p$) are 500000 and 4, respectively. The electron bunch is divided into 1209 slices ($=L$) with the slice positions of $s$ = -9.99382e-07,-9.97437e-07,..., and the radiation field is evaluated at 128 ($=M$) x 128 ($=N$) transverse grid points, with the intervals of $(\Delta x,\Delta y,\Delta x',\Delta y')=$(2.13045e-06,1.23744e-06,4.75329e-07,8.18358e-07).
Radiation Profiles
The complex amplitude of radiation $E(z,s,\mathbf{r})$ is saved in file(s) named as "*-h.fld", with h being the harmonic number. For computational efficiency, the angular representation $\mathscr{E}$ is saved instead of $E$, which is defined by the spatial Fourier transform, \[\mathscr{E}(z,s,\mathbf{r}')=\int E(z,s,\mathbf{r})\exp(-i\kappa\mathbf{r}'\cdot\mathbf{r})d\mathbf{r},\] where $\kappa=2\pi/\lambda$ is the wavenumber of radiation, and $\mathbf{r}'=(x',y')$ is the angular coordinate defined as the Fourier conjugate of the transverse coordinate $\mathbf{r}$. The discrete data set of $A_{klmn}=\mathscr{E}(z_k,s_l,\mathbf{r}'_{m,n}=x'_m,y'_n)$ is saved in the format as follows.
\[ \begin{array}{ccccccc} A_{0000,re} & A_{0000,im} & A_{0001,re} & A_{0001,im} & \ldots & A_{000N',re} & A_{000N',im} \\ A_{0010,re} & A_{0010,im} & A_{0011,re} & A_{0011,im} & \ldots & A_{00M'N',re} & A_{00M'N',im} \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ A_{0100,re} & A_{0100,im} & A_{0101,re} & A_{0101,im} & \ldots & A_{010N',re} & A_{010N',im} \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ A_{1000,re} & A_{1000,im} & A_{1001,re} & A_{1001,im} & \ldots & A_{100N',re} & A_{100N',im} \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ A_{K'L'M'0,re} & A_{K'L'M'0,im} & A_{K'L'M'1,re} & A_{K'L'M'1,im} & \ldots & A_{K'L'M'N',re} & A_{K'L'M'N',im} \\ \end{array} \]
where subscripts re and im mean the real and imaginary parts and $K'=K-1$ being the maximum index for $z_k$, etc. Then, we have $2KLMN$ float numbers in total in this file.
The relation between indices $(k,l,m,n)$ and real coordinates $(z,s,x',y')$ are given in the output file (*.json) in the former section. Note that the transverse grid points $x,y,x',y'$ are indexed by a common FFT algorithm, and are not as straightforward as $z$ and $s$. Namely, \[x_m=\left\{ \begin{array}{ll} m\Delta x&;\: m\leq M/2 \\ (m-M)\Delta x&;\: m> M/2 \end{array} \right.\] and a similar expression for $y,x',y'$.
The spatial profile of the complex amplitude, $E(\mathbf{r})$, can be restored by Fourier transforming $\mathscr{E}(\mathbf{r}')$, with which $E(\mathbf{r})$ is discretely given at transverse grid points $x_m$ and $y_m$. Note that we have a well-known relation \[\Delta x' = \frac{\lambda}{M\Delta x},\] and thus $\Delta x',\Delta y'$ should be divided by the harmonic number h for higher harmonics.
If a special (neither linear nor helical) undulator is selected, the radiation field cannot be given by a scalar, and the file format is slghtly different. To be specific, the horizontal ($E_x$) and vertical ($E_y$) components are alternately saved at each longitudinal step, and the format looks as follows.
\begin{eqnarray} \left. \begin{array}{ccccccc} A_{x,0000,re} & A_{x,0000,im} & A_{x,0001,re} & A_{x,0001,im} & \ldots & A_{x,000N',re} & A_{x,000N',im} \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ A_{x,0L'M'0,re} & A_{x,0L'M'0,im} & A_{x,0L'M'1,re} & A_{x,0L'M'1,im} & \ldots & A_{x,0L'M'N',re} & A_{x,0L'M'N',im} \\ \end{array} \right\} &\Rightarrow& \mbox{(0)} \\ \left. \begin{array}{ccccccc} A_{y,0000,re} & A_{y,0000,im} & A_{y,0001,re} & A_{y,0001,im} & \ldots & A_{y,000N',re} & A_{y,000N',im} \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ A_{y,0L'M'0,re} & A_{y,0L'M'0,im} & A_{y,0L'M'1,re} & A_{y,0L'M'1,im} & \ldots & A_{y,0L'M'N',re} & A_{y,0L'M'N',im} \\ \end{array} \right\} &\Rightarrow& \mbox{(1)} \\ \left. \begin{array}{ccccccc} A_{x,1000,re} & A_{x,1000,im} & A_{x,1001,re} & A_{x,1001,im} & \ldots & A_{x,100N',re} & A_{x,100N',im} \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ A_{y,K'L'M'0,re} & A_{y,K'L'M'0,im} & A_{y,K'L'M'1,re} & A_{y,K'L'M'1,im} & \ldots & A_{y,K'L'M'N',re} & A_{y,K'L'M'N',im} \\ \end{array} \right\} &\Rightarrow& \mbox{(2-K')} \\ \end{eqnarray}
The section (0) indicted by a curly brace contains $2LMN$ float numbers, representing the horizontal components ($E_x$) over the whole slices and transverse grid points at the 0th step, and the section (1) represents the vertical components at the same step. This arrangement continues until the end of the longitudinal step ($K'$-th step).
Partile Distribution
The distribution of macroparticles is saved in a file named as "*.par". As explained in Structure of a Beamlet, the electron beam in SIMPLEX is composed of $N_b$ beamlets, each of which is defined by $4+2N_p$ coordinate variables. The first 4 define the transverse position ($x,y$) and angle ($x',y'$) of each beamlet, while the latter $2N_p$ define the energy ($\Delta\gamma/\gamma$) and longitudinal position ($s$) of each macroparticle. In SIMPLEX, the tansverse coordinate of each beamlet is simply evaluated by the transfer matrix formalism, and can be easily evaluated at any longitudinal steps once the initial values are given. For example, we have \[ \left[ \begin{array}{c} x(z) \\ x'(z) \\ \eta \end{array} \right] = \left[ \begin{array}{ccc} C_x(z) & S_x(z) & D_x(z) \\ C'_x(z) & S'_x(z) & D'_x(z) \\ 0 & 0 & 1 \end{array} \right] \left[ \begin{array}{c} x(0) \\ x'(0) \\ \eta \end{array} \right] \] with $\eta\equiv\Delta\gamma/\gamma$ being relative energy, and a similar expression for $y$ and $y'$. In contrast, the energy and longitudinal position of each macroparticle are numerically evaluated by solving the relevant equations and are saved at each step. As a result, the particle data file has a data format as follows.
\[\begin{eqnarray} \begin{array}{ccccccccccc} x_0 & y_0 & x'_0 & y'_0 & x_1 & \ldots & x'_{N_b'-1} & y'_{N_b'-1} & x_{N_b'} & y_{N_b'} & x'_{N_b'} & y'_{N_b'} \\ \end{array} \:\:\:\:\:\: &\Rightarrow& (0) \\ \left. \begin{array}{ccccccccccc} C_{x,0} & S_{x,0} & D_{x,0} & C'_{x,0} & S'_{x,0} & D'_{x,0} & C_{y,0} & S_{y,0} & D_{y,0} & C'_{y,0} & S'_{y,0} & D'_{y,0} \\ & & & & & \vdots & \vdots & & & & & \\ C_{x,K'} & S_{x,K'} & D_{x,K'} & C'_{x,K'} & S'_{x,K'} & D'_{x,K'} & C_{y,K'} & S_{y,K'} & D_{y,K'} & C'_{y,K'} & S'_{y,K'} & D'_{y,K'} \\ \end{array} \right\} &\Rightarrow& (1) \\ \left. \begin{array}{ccccccccccc} s_{000} & \eta_{000} & s_{001} & \eta_{001} & \ldots & s_{00N_p'} & \eta_{00N_p'} & s_{010} & \eta_{010} & \ldots & s_{0N_b'N_p'} & \eta_{0N_b'N_p'} \\ s_{100} & \eta_{100} & s_{101} & \eta_{101} & \ldots & s_{10N_p'} & \eta_{10N_p'} & s_{110} & \eta_{110} & \ldots & s_{1N_b'N_p'} & \eta_{1N_b'N_p'} \\ & & & & & \vdots & \vdots & & & & & \\ s_{K'00} & \eta_{K'00} & s_{K'01} & \eta_{K'01} & \ldots & s_{K'0N_p'} & \eta_{K'0N_p'} & s_{K'10} & \eta_{K'10} & \ldots & s_{K'N_b'N_p'} & \eta_{K'N_b'N_p'} \\ \end{array} \right\} &\Rightarrow& (2) \end{eqnarray} \]
where $x_j, y_j, x_j', y_j'$ denote the transverse coordinate of the $j$-th beamlet, $C_{x,k}, S_{x,k}, ...$ denote the transfer matrix elements at the $k$-th step, and $s_{kji}$ and $\eta_{kji}$ denotes the longitudinal position and relative energy of the $i$-th particle in the $j$-th beamlet at the $k$-th step. As shown above, the data is composed of 3 sections. The section (0) contains $4N_b$ numbers defining the initial transverse coodrinate of the beamlets, section (1) contains $12K$ numbers to specify the transfer matrix, and section (2) contains $KN_bN_p$ numbers to describe the motion of macroparticles in the energy-time phase space while the electron bunch travels along the undulator.
Standalone Mode
Besides the desktop application as described above, the solver of SIMPLEX can be run in a standalone mode. Note that in the followings, [simplex_home] refers to the directory where SIMPLEX has been installed.
When [Run]-[Start Simulation] command is executed, the SIMPLEX GUI creates an input parameter file ("*.json") and invokes the solver ("simplex_solver" or "simplex_solver_nompi" depending on whether the parallel computing option is enabled or not) located in the same directory as the main GUI program, with the input file as an argument. This means that SIMPLEX (solver) can be run without the GUI, if the input file is prepared by an alternative method, and a batch process will be possible. To do so, prepare the input file according to the descriptions in Input Format and run the solver as follows
[simplex_home]/simplex_solver_nompi -f [input file]to start a simulation without parallel computing, or
mpiexec -n 4 [simplex_home]/simplex_solver_nompi -f [input file]with parallel computing (with 4 MPI processes in this case).
It should be noted that the names of the parameters and options ("key" of the object) should be totally correct, including the units and inserted space characters. In addition, the number of parameters and options actually needed for a specific simulation depend on its condition. To avoid possible errors and complexity in preparing the input file, it is recommended to create a "master input file" specific to the desired simulation condition, by running [Run]-[Export Simulation Settings]. Then, just modify the values of desired parameters to prepare the input file.
Note that this usage has not been officially supported before ver. 2.1, simply because the input file format was original and difficult to read.
Python User Interface
To support the expanding users of python language, SIMPLEX has started official support for python, since ver. 3.1. To make use of this function, python version 3.8 or later is needed, and the user is requested to install a python package simplex-ui (SIMPLEX User Interface). Refer to the instructions for details of simplex-ui.
References
- T. Tanaka, "SIMPLEX: SIMulator and Postprocessor for free electron Laser EXperiments," J. Synchrotron Radiation 22, 1319 (2015)
- R. Bonifacio, C. Pellegrini and L. M. Narducci, "Collective instabilities and high-gain regime in a free electron laser," Optics Communications, 50 (1984) 373
- E. L. Saldin, E. A. Schneidmiller and M. V. Yurkov, The Physics of Free Electron Lasers, Springer, 2000
- M. Xie, "Exact and variational solutions of 3D eigenmodes in high gain FELs," Nucl. Instrum. Meth. A445 (2000) 59
- K. L. F. Bane and G. Stupakov, "Resistive wall wakefield in the LCLS undulator beam pipe," SLAC-PUB-10707 (2004)
- G. V. Stupakov, "Impedance of small obstacles and rough surfaces," Phys. Rev. ST-AB, 1 (1998) 064401
- G. V. Stupakov, "Surface roughness impedance," Proc. ICFA Workshop on Phys. Sci. X-ray FEL, 2000
- G. V. Stupakov, "Surface impedance and synchronous modes," Workshop on Instabilities of High Intensity Hadron Beams in Rings, 1999
- M. Venturini, "Models of longitudinal space-charge impedance for microbunching instability," Phys. Rev. ST-AB 11, 034401 (2008)
- J. Feldhaus et al., "Possible application of X-ray optical elements for reducing the spectral bandwidth of an X-ray SASE FEL," Optics Comm., 140 (1997) 341
- G. Geloni, V. Kocharyan and E. Saldin, "A novel self-seeding scheme for hard x-ray FELs," J. Mod. Optics, 58 (2011) 1391
- T. Tanaka "Accelerating the convergence of free electron laser simulations by retrieving a spatially-coherent component of microbunching," arXiv:2310.20197
- W. M. Fawley, "Algorithm for loading shot noise microbunching in multidimensional free-electron laser simulation codes," Phys. Rev. ST-AB, 5 (2002) 070701
- R. Walker, "Interference effects in undulator and wiggler radiation sources", Nucl. Instrum. Methods Phys. Res., Sect. A 335, 328 (1993)
Acknowledgements
This software includes the work that is distributed in the Apache License 2.0, and relies on a number of libraries as summarized below, which are gratefully appreciated.
- Node.js: Javascript runtime environment to run Javascript code without a web browser (https://nodejs.org/en/).
- Tauri: toolkit to build a desktop application with Javascript, HTML and CSS (https://www.electronjs.org/)
- Plotly.js: Javascript graphing library to facilitate data visualization (https://nodejs.org/en/)
- mathjax: Javascript library to display mathematical formulas in HTML documents (https://www.mathjax.org/)
- picojson: JSON parser for C++ language (https://github.com/kazuho/picojson)
- General Purpose FFT Package (https://www.kurims.kyoto-u.ac.jp/~ooura/fft.html)